Quickstart guide
About
R wrapper for iNaturalist APIs for accessing the observations. The detailed documentation of API is available on iNaturalist website and is part of our larger species occurence searching packages SPOCC.
Get observations
Searching
Fuzzy search
You can search for observations by either common or latin name. It will search the entire iNaturalist entry, so the search below will return all entries that mention Monarch butterflies, not just entries for Monarchs.
library(rinat)
butterflies <- get_inat_obs(query = "Monarch Butterfly")
Another use for a fuzzy search is searching for a common name or habitat, e.g. searching for all observations that might happen in a vernal pool. We can then see all the species names found.
vp_obs <- get_inat_obs(query = "vernal pool")
head(vp_obs$species_guess)
## [1] "Bolander's Sunflower" "American royal fern"
## [3] "" ""
## [5] "Wood Frog" "Twelve-spotted Skimmer"
Taxon query
To return only records for a specific species or taxonomic group, use the taxon option.
## Return just observations in the family Plecoptera
stone_flies <- get_inat_obs(taxon_name = "Plecoptera", year = 2010)
## Return just Monarch Butterfly records
just_butterflies <- get_inat_obs(taxon_name = "Danaus plexippus")
Bounding box search
You can also search within a bounding box by giving a simple set of coordinates.
## Search by area
bounds <- c(38.44047, -125, 40.86652, -121.837)
deer <- get_inat_obs(query = "Mule Deer", bounds = bounds)
Other functions
Get information and observations by project
You can get all the observations for a project if you know it’s ID or name as an iNaturalist slug.
## Just get info about a project
vt_crows <- get_inat_obs_project("crows-in-vermont", type = "info", raw = FALSE)
## 164 Records
## Now get all the observations for that project
vt_crows_obs <- get_inat_obs_project(vt_crows$id, type = "observations")
## 164 Records
## 0-200
Get observation details
Detailed information about a specific observation can be retrieved by observation ID. The easiest way to get the ID is from a more general search.
m_obs <- get_inat_obs(query = "Monarch Butterfly")
head(get_inat_obs_id(m_obs$id[1]))
## $id
## [1] 28657566
##
## $observed_on
## [1] "2019-07-09"
##
## $description
## NULL
##
## $latitude
## [1] "42.0774822254"
##
## $longitude
## [1] "-83.1035997957"
##
## $map_scale
## NULL
Get all observations by user
If you just want all the observations by a user you can download all their observations by user ID. A word of warning though, this can be quite large (easily into the 1000’s).
m_obs <- get_inat_obs(query = "Monarch Butterfly")
head(get_inat_obs_user(as.character(m_obs$user_login[1]), maxresults = 20))[,1:5]
## scientific_name datetime description place_guess
## 1 Phidippus audax 2019-06-07 16:44:09 -0400 Ontario, CA
## 2 Danaus plexippus 2019-07-09 11:16:54 -0400 Ontario, CA
## 3 Danaus plexippus 2019-07-11 16:24:13 -0400 Ontario, CA
## 4 Oecanthus niveus 2019-07-11 16:20:46 -0400 Ontario, CA
## 5 Megachile mendica 2019-07-11 16:22:04 -0400 Ontario, CA
## 6 Lestes rectangularis 2019-07-09 17:01:12 -0400 Ontario, CA
## latitude
## 1 42.00755
## 2 42.07748
## 3 42.01950
## 4 42.19113
## 5 42.04144
## 6 42.10027
Stats by taxa
Basic statistics are available for taxa counts by date, date range, place ID (numeric ID), or user ID (string).
## By date
counts <- get_inat_taxon_stats(date = "2010-06-14")
print(counts$total)
## [1] 383
print(counts$species_counts[1:5,])
## count taxon.id taxon.name taxon.rank taxon.rank_level
## 1 5 41708 Phoca vitulina species 10
## 2 5 57495 Melitaea cinxia species 10
## 3 3 48662 Danaus plexippus species 10
## 4 3 52766 Megaphasma denticrus species 10
## 5 3 56057 Leucanthemum vulgare species 10
## taxon.default_name.id taxon.default_name.name
## 1 67153 Harbor Seal
## 2 91278 Glanville Fritillary
## 3 586653 Monarch
## 4 83649 Giant Walkingstick
## 5 924783 oxeye daisy
## taxon.default_name.is_valid taxon.default_name.lexicon
## 1 TRUE English
## 2 TRUE English
## 3 TRUE English
## 4 TRUE English
## 5 TRUE English
## taxon.default_name.taxon_id taxon.default_name.created_at
## 1 41708 2008-03-19T00:35:25.000Z
## 2 57495 2010-03-17T06:57:02.000Z
## 3 48662 2014-09-11T06:50:44.683Z
## 4 52766 2009-08-24T02:06:09.000Z
## 5 56057 2017-06-08T19:00:12.460Z
## taxon.default_name.updated_at taxon.default_name.creator_id
## 1 2019-03-25T06:53:47.770Z NA
## 2 2010-03-17T06:57:02.000Z NA
## 3 2015-07-22T12:00:15.313Z NA
## 4 2009-08-24T02:06:09.000Z NA
## 5 2018-01-10T00:03:27.724Z 498994
## taxon.default_name.position
## 1 0
## 2 0
## 3 1
## 4 0
## 5 17
## taxon.image_url
## 1 https://static.inaturalist.org/photos/3238907/square.jpg?1459150554
## 2 https://static.inaturalist.org/photos/5403994/square.jpg?1545659971
## 3 https://static.inaturalist.org/photos/1477/square.jpg?1545368454
## 4 https://static.inaturalist.org/photos/333073/square.jpg?1444606035
## 5 https://static.inaturalist.org/photos/361971/square.jpg?1444643087
## taxon.iconic_taxon_name taxon.conservation_status_name
## 1 Mammalia least_concern
## 2 Insecta <NA>
## 3 Insecta <NA>
## 4 Insecta <NA>
## 5 Plantae <NA>
print(counts$rank_counts)
## $species
## [1] 325
##
## $genus
## [1] 19
##
## $subspecies
## [1] 18
##
## $family
## [1] 6
##
## $order
## [1] 3
##
## $variety
## [1] 3
##
## $class
## [1] 2
##
## $kingdom
## [1] 2
##
## $phylum
## [1] 1
##
## $subclass
## [1] 1
##
## $subfamily
## [1] 1
##
## $superfamily
## [1] 1
##
## $tribe
## [1] 1
Stats by user
Similar statistics can be gotten for users. The same input parameters can be used, but results are the top five users by species count and observation count.
## By date
counts <- get_inat_user_stats(date = "2010-06-14")
print(counts$total)
## [1] 155
print(counts$most_observations[1:10,])
## count user.id user.login user.name
## 1 53 811118 sandbankspp
## 2 24 761669 kathleenfspicer <NA>
## 3 20 922078 hakai470 ES470: Hakai Institute
## 4 12 357375 richardling Richard Ling
## 5 12 522214 tdavenport Tripp Davenport
## 6 10 9706 greglasley Greg Lasley
## 7 9 1422334 adriao <NA>
## 8 8 109098 leannewallis Leanne Wallis
## 9 8 362446 pwdeacon Pat Deacon
## 10 8 677594 nakarb Robin Bad
## user.user_icon_url
## 1 https://static.inaturalist.org/attachments/users/icons/811118/thumb.jpeg?1535236899
## 2 <NA>
## 3 <NA>
## 4 https://static.inaturalist.org/attachments/users/icons/357375/thumb.jpg?1484462740
## 5 https://static.inaturalist.org/attachments/users/icons/522214/thumb.jpg?1558671195
## 6 https://static.inaturalist.org/attachments/users/icons/9706/thumb.jpg?1533329961
## 7 <NA>
## 8 https://static.inaturalist.org/attachments/users/icons/109098/thumb.jpg?1475547611
## 9 https://static.inaturalist.org/attachments/users/icons/362446/thumb.jpg?1546674469
## 10 <NA>
print(counts$most_species[1:10,])
## count user.id user.login user.name
## 1 47 811118 sandbankspp
## 2 22 761669 kathleenfspicer <NA>
## 3 14 922078 hakai470 ES470: Hakai Institute
## 4 10 9706 greglasley Greg Lasley
## 5 10 357375 richardling Richard Ling
## 6 9 522214 tdavenport Tripp Davenport
## 7 9 1422334 adriao <NA>
## 8 8 362446 pwdeacon Pat Deacon
## 9 8 1519122 steven307 Steven Joyner
## 10 7 109098 leannewallis Leanne Wallis
## user.user_icon_url
## 1 https://static.inaturalist.org/attachments/users/icons/811118/thumb.jpeg?1535236899
## 2 <NA>
## 3 <NA>
## 4 https://static.inaturalist.org/attachments/users/icons/9706/thumb.jpg?1533329961
## 5 https://static.inaturalist.org/attachments/users/icons/357375/thumb.jpg?1484462740
## 6 https://static.inaturalist.org/attachments/users/icons/522214/thumb.jpg?1558671195
## 7 <NA>
## 8 https://static.inaturalist.org/attachments/users/icons/362446/thumb.jpg?1546674469
## 9 https://static.inaturalist.org/attachments/users/icons/1519122/thumb.jpeg?1555274551
## 10 https://static.inaturalist.org/attachments/users/icons/109098/thumb.jpg?1475547611
## By place_ID
vt_crows <- get_inat_obs_project("crows-in-vermont", type = "info", raw = FALSE)
## 164 Records
place_counts <- get_inat_user_stats(place = vt_crows$place_id)
print(place_counts$total)
## [1] 5536
print(place_counts$most_observations[1:10,])
## count user.id user.login user.name
## 1 41694 12158 erikamitchell Erika Mitchell
## 2 28614 2179 charlie Charlie Hohn
## 3 13096 12610 susanelliott Susan Elliott
## 4 7887 12045 larry522 Larry Clarfeld
## 5 7680 12036 zaccota Zac Cota
## 6 6440 317 kpmcfarland Kent McFarland
## 7 6165 28921 rwp84 roy pilcher
## 8 6144 108365 judywelna
## 9 5268 11792 kylejones Kyle Jones
## 10 5162 6624 joannerusso
## user.user_icon_url
## 1 https://static.inaturalist.org/attachments/users/icons/12158/thumb.jpg?1558486310
## 2 https://static.inaturalist.org/attachments/users/icons/2179/thumb.jpg?1475528361
## 3 https://static.inaturalist.org/attachments/users/icons/12610/thumb.jpg?1475533475
## 4 https://static.inaturalist.org/attachments/users/icons/12045/thumb.jpg?1475533238
## 5 https://static.inaturalist.org/attachments/users/icons/12036/thumb.jpg?1475533232
## 6 https://static.inaturalist.org/attachments/users/icons/317/thumb.jpg?1475527502
## 7 https://static.inaturalist.org/attachments/users/icons/28921/thumb.jpg?1475542431
## 8 https://static.inaturalist.org/attachments/users/icons/108365/thumb.jpg?1475547470
## 9 https://static.inaturalist.org/attachments/users/icons/11792/thumb.jpg?1475533125
## 10 https://static.inaturalist.org/attachments/users/icons/6624/thumb.jpeg?1562532360
print(place_counts$most_species[1:10,])
## count user.id user.login user.name
## 1 2444 12158 erikamitchell Erika Mitchell
## 2 2054 12045 larry522 Larry Clarfeld
## 3 2051 12610 susanelliott Susan Elliott
## 4 1703 2179 charlie Charlie Hohn
## 5 1509 6624 joannerusso
## 6 1489 1088797 montpelierbioblitz1 Montpelier BioBlitz
## 7 1453 11792 kylejones Kyle Jones
## 8 1289 317 kpmcfarland Kent McFarland
## 9 1261 108365 judywelna
## 10 1086 12049 gaudettelaura Laura Gaudette
## user.user_icon_url
## 1 https://static.inaturalist.org/attachments/users/icons/12158/thumb.jpg?1558486310
## 2 https://static.inaturalist.org/attachments/users/icons/12045/thumb.jpg?1475533238
## 3 https://static.inaturalist.org/attachments/users/icons/12610/thumb.jpg?1475533475
## 4 https://static.inaturalist.org/attachments/users/icons/2179/thumb.jpg?1475528361
## 5 https://static.inaturalist.org/attachments/users/icons/6624/thumb.jpeg?1562532360
## 6 <NA>
## 7 https://static.inaturalist.org/attachments/users/icons/11792/thumb.jpg?1475533125
## 8 https://static.inaturalist.org/attachments/users/icons/317/thumb.jpg?1475527502
## 9 https://static.inaturalist.org/attachments/users/icons/108365/thumb.jpg?1475547470
## 10 https://static.inaturalist.org/attachments/users/icons/12049/thumb.jpg?1475533241
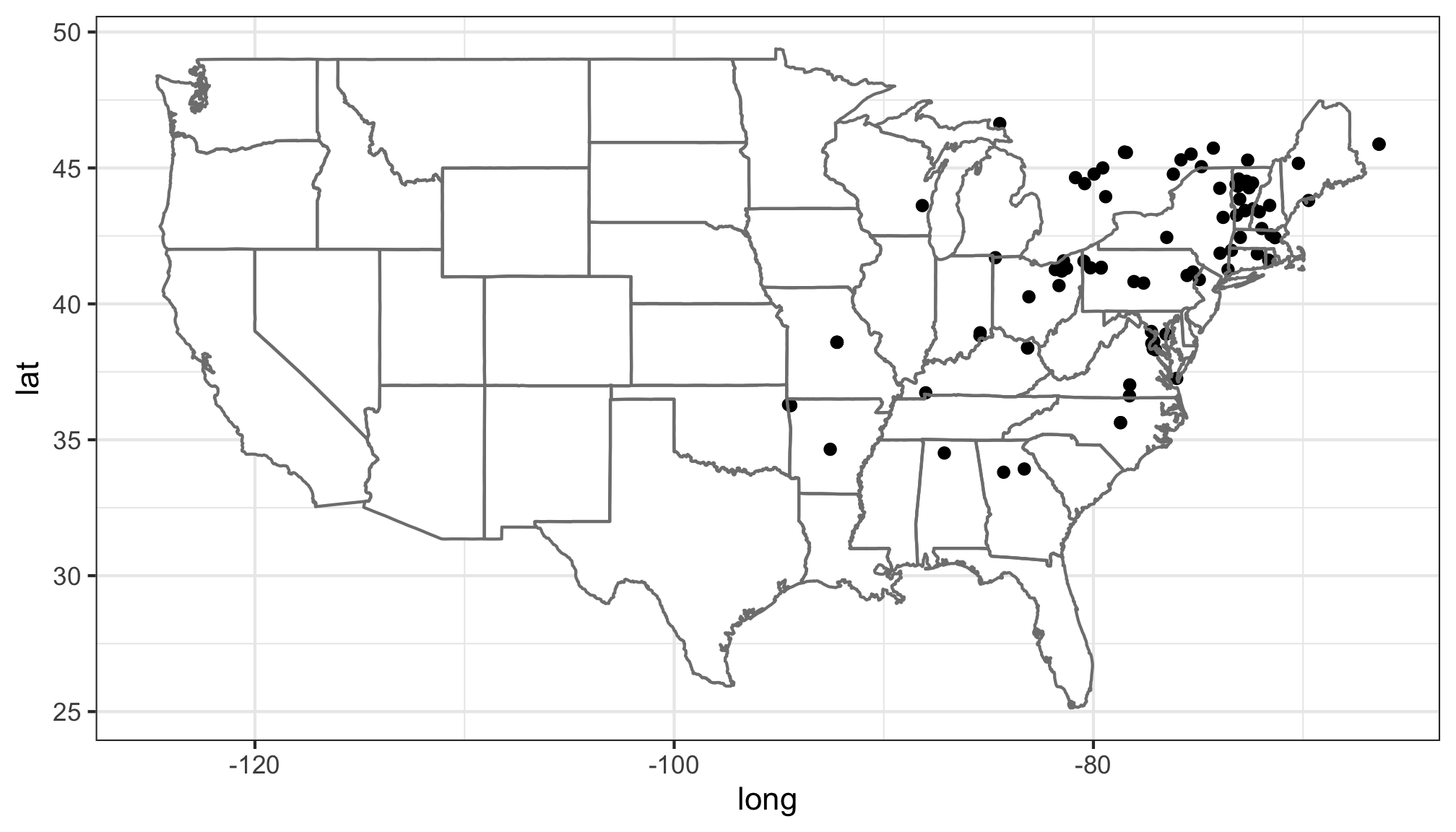
Mapping
Basic maps can be created as well to quickly visualize search results.
Maps can either be plotted automatically plot = TRUE or simply return
a ggplot2 object with plot = FALSE. This works well with single
species data, but more complicated plots are best made from scratch.
library(ggplot2)
## Map salamanders in the genuse Ambystoma
m_obs <- get_inat_obs(taxon_name = "Ambystoma maculatum")
salamander_map <- inat_map(m_obs, plot = FALSE)
### Now we can modify the returned map
salamander_map + borders("state") + theme_bw()
## Warning: Removed 2 rows containing missing values (geom_point).