This is code to reproduce key results and figures from the article: Large Scale Gene Set Ranking for Survival-Related Gene Sets.
The code was tested on Ubuntu 20.04.4 LTS and MacOS 13.1. With python versions 3.9.15 and 3.10.8.
Follow these steps to prepare the environment:
- Clone the repository
git clone https://github.com/MartinSpendl/survival-geneset-ranking
cd survival-geneset-ranking- Install the required packages
# using pip in a virtual environment
pip install -r requirements.txt
# using Conda
conda create --name <env_name> --file requirements.txt
conda activate <env_name>Data used for the analysis is publically accessible.
Download repository and run all 4 notebooks: https://github.com/JakaKokosar/tcga-data.git
Download genesets from the MSigDB by signing in.
Save them to the data/genesets directory.
We used:
- Hallmark genesets:
h.all.v2023.1.Hs.symbols.gmt - GO-Biological Process:
c5.go.bp.v2023.1.Hs.symbols.gmt
Firstly, run both scipts in the /scipts directory:
python calculate_ssGSEA.py
python calculate_statistical_test.pySecondly, run notebooks in the /notebooks directory.
Figures from the notebooks are stored in the /figures directory.
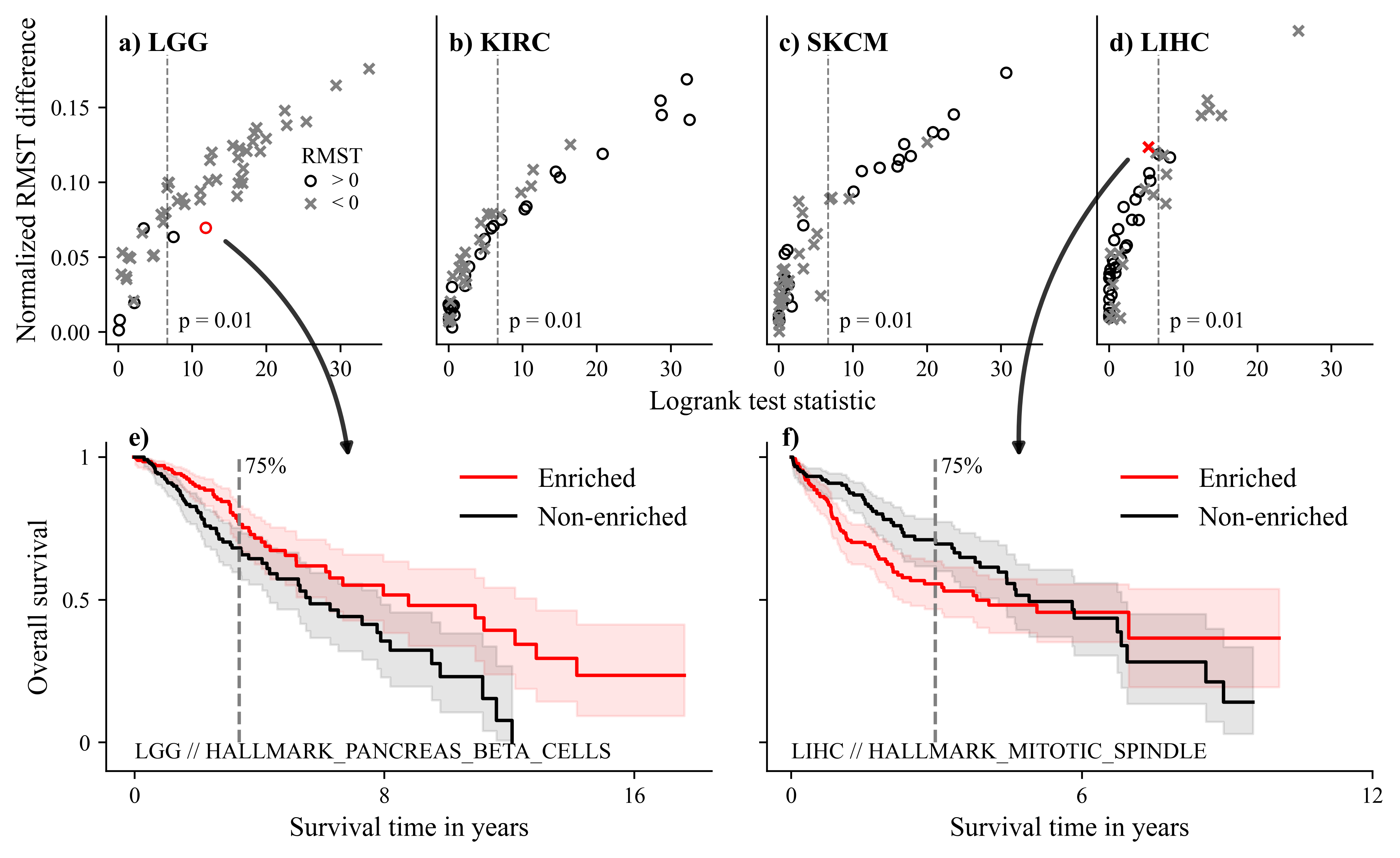
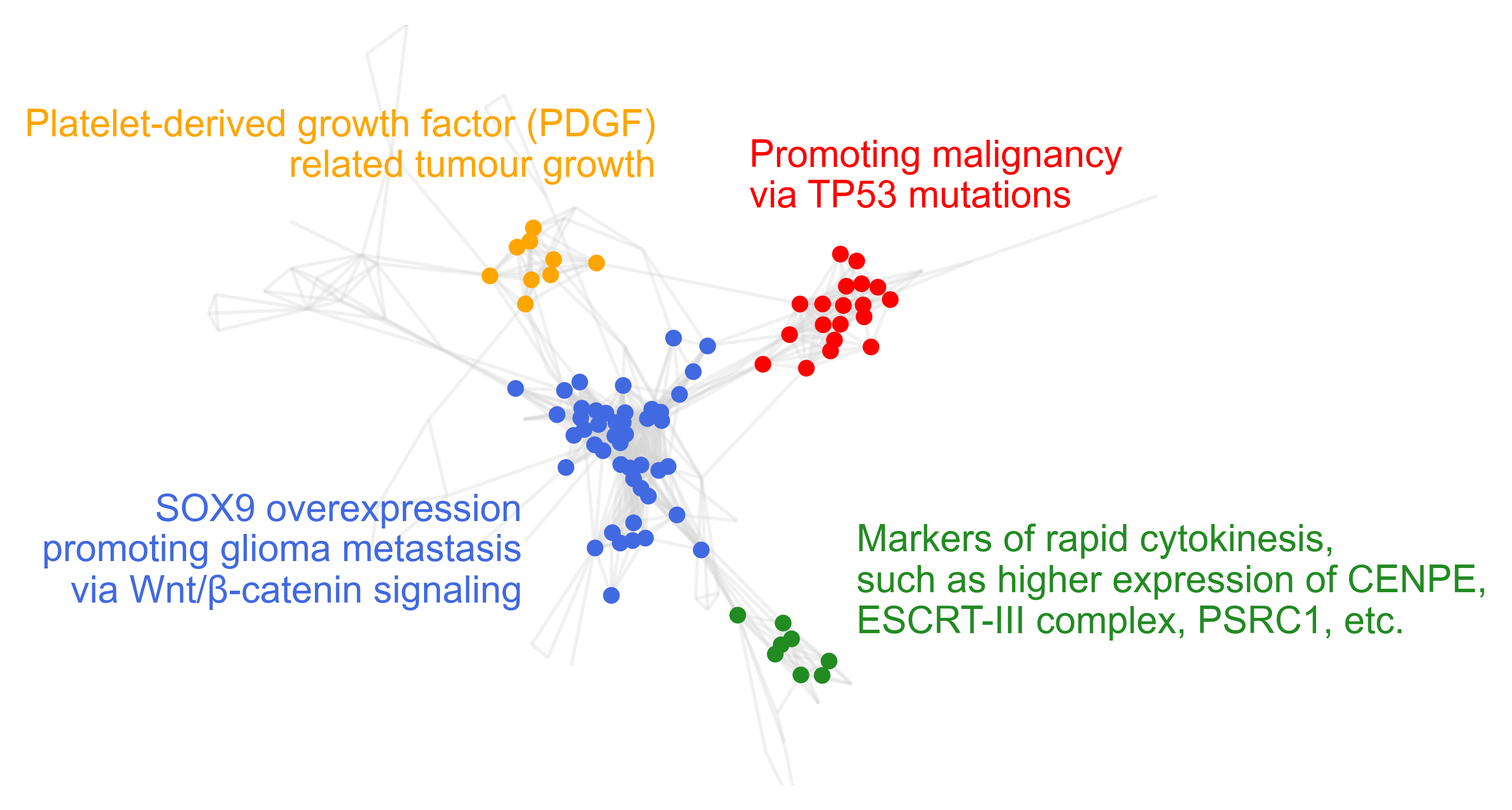
Cluters of survival-related genesets from GO gene sets on the TCGA-KIRC dataset.