Repository for projects of the ETH Zürich course "Machine Learning for Health Care (Spring 2020)" (lecture page).
Authors :
Han Bai
Nora Moser
Martin Tschechne (martints@ethz.ch)
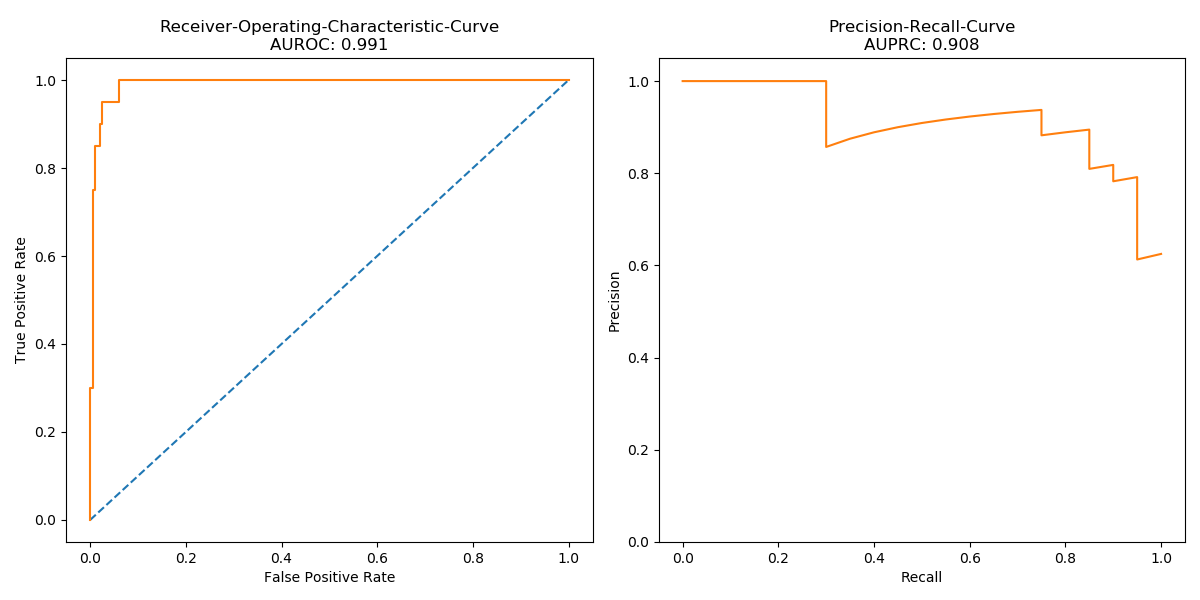
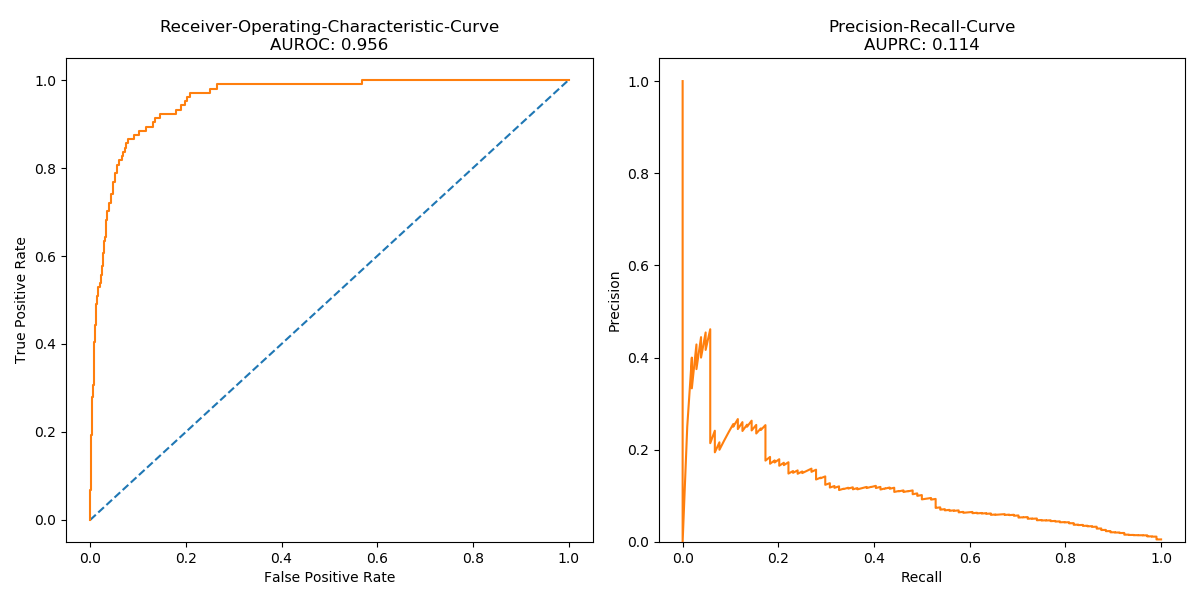
Classifying ECG signals of the MIT-BIH Arrhythmia Dataset and the PTB Diagnostic ECG Database by Recurrent Neural Networks and make use of Transfer Learning techniques in order to improve predictive performance.
Results
| Models | MIT-BIH* | PTBDB* | PTBDB° | PTBDB† |
|---|---|---|---|---|
| LSTM + FC | F1: 0.184 Acc: 0.823 |
F1: 0.787 Acc: 0.776 AUROC: 0.808 AUPRC: 0.934 |
F1: 0.419 Acc: 0.722 AUROC: 0.5 AUPRC: 0.861 |
F1: 0.371 Acc: 0.565 AUROC: 0.397 AUPRC: 0.805 |
| CNN + LSTM + FC | F1: 0.868 Acc: 0.971 |
F1: 0.940 Acc: 0.951 AUROC: 0.947 AUPRC: 0.982 |
F1: 0.988 Acc: 0.990 AUROC: 0.988 AUPRC: 0.996 |
F1: 0.992 Acc: 0.994 AUROC: 0.990 AUPRC: 0.996 |
| LSTM + XGB‡ | F1: 0.875 Acc: 0.976 |
F1: 0.971 Acc: 0.977 AUROC: 0.968 AUPRC: 0.988 |
F1: 0.963 Acc: 0.970 AUROC: 0.955 AUPRC: 0.983 |
- |
| CNN + LSTM + XGB‡ | F1: 0.916 Acc: 0.985 |
F1: 0.983 Acc: 0.986 AUROC: 0.980 AUPRC: 0.993 |
F1: 0.981 Acc: 0.990 AUROC: 0.977 AUPRC: 0.991 |
- |
| XGB | F1: 0.896 Acc: 0.979 |
F1: 0.970 Acc: 0.976 AUROC: 0.966 AUPRC: 0.987 |
- | - |
| Kachuee, et al.[1] | Acc: 0.934 | - | F1: 0.951 Acc: 0.959 |
- |
| Baseline[2] | F1: 0.915 Acc: 0.985 |
F1: 0.988 Acc: 0.983 |
F1: 0.969 Acc: 0.956 |
F1: 0.994 Acc: 0.992 |
* Only trained on this dataset
° Transfer Learning, pre-trained model trained on MIT-BIH, retrained with frozen base layers
† Transfer Learning, pre-trained model trained on MIT-BIH, retrained with unfrozen base layers
‡ Base layers always frozen to train XGBoost
Visualization of learned embeddings
| t-SNE | UMAP | PCA | |
|---|---|---|---|
| MIT-BIH | 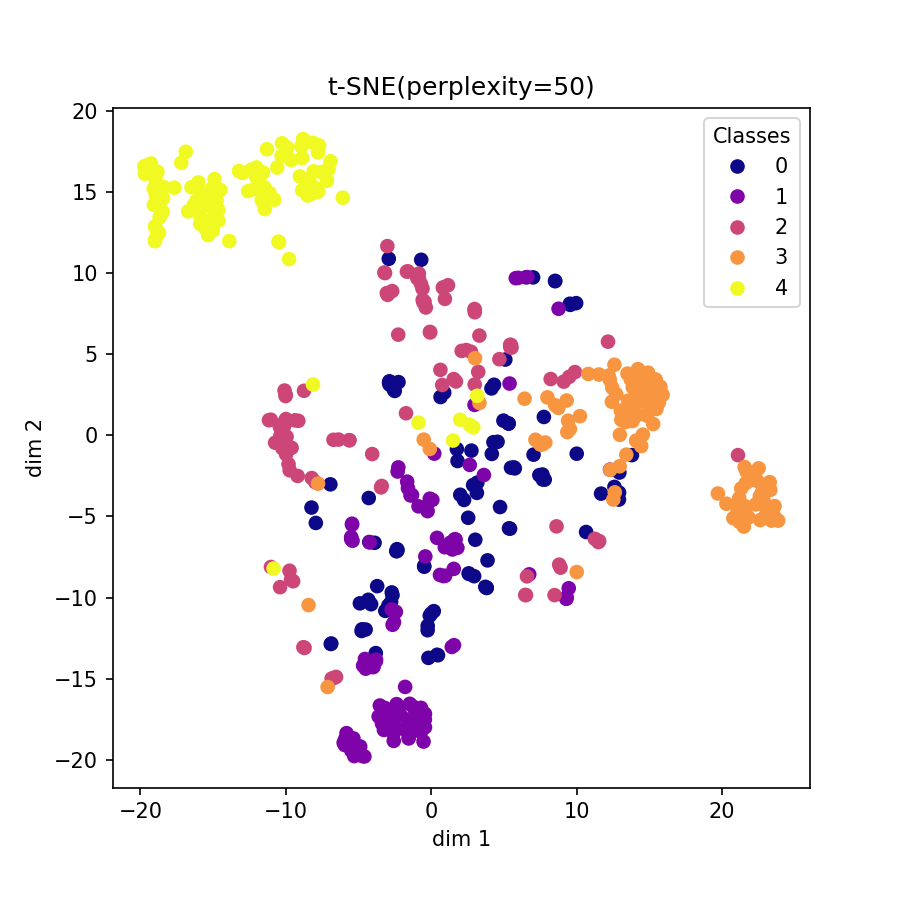 |
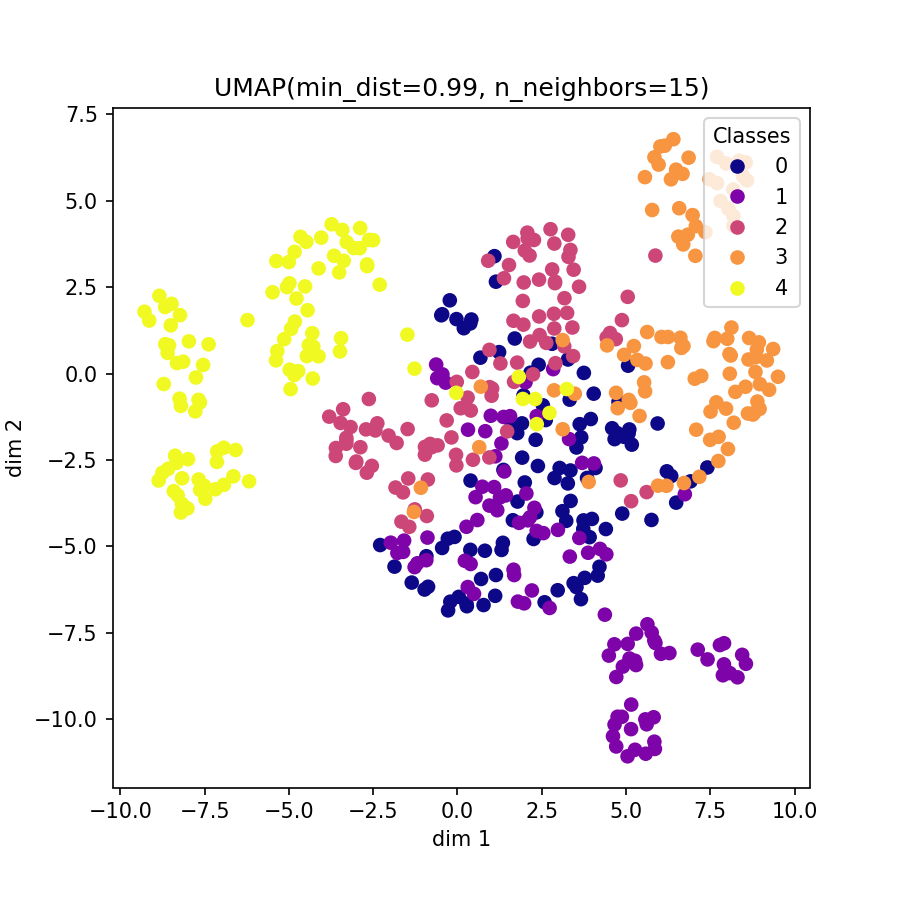 |
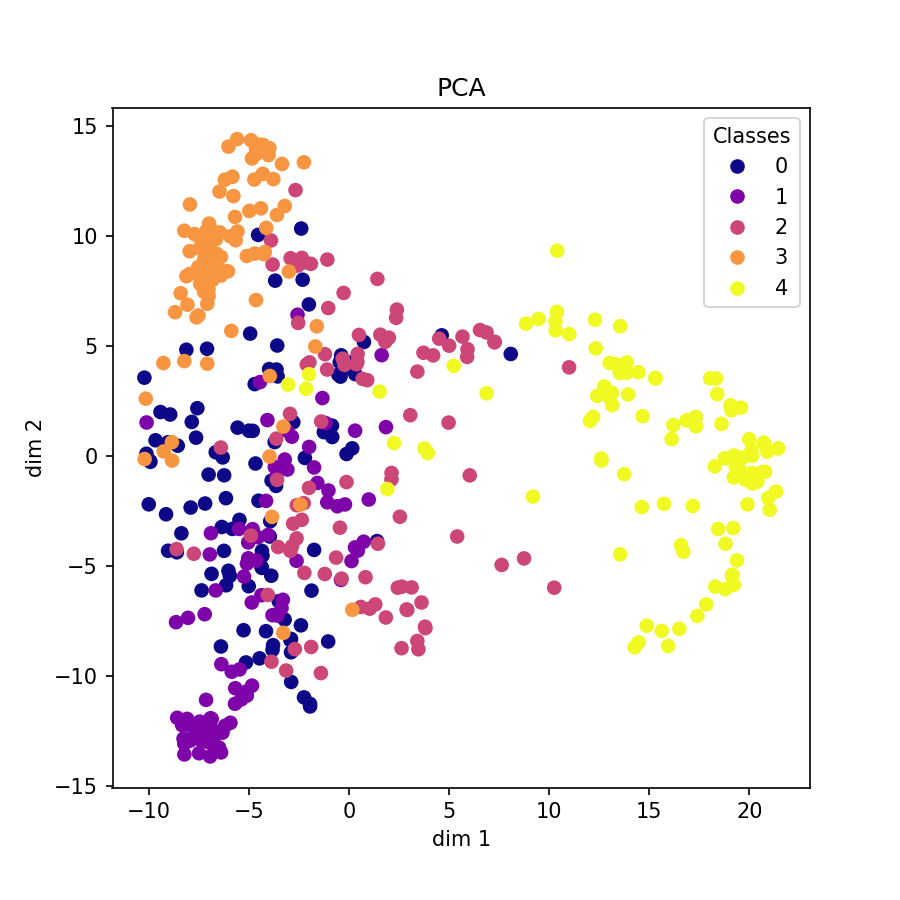 |
| PTBDB | 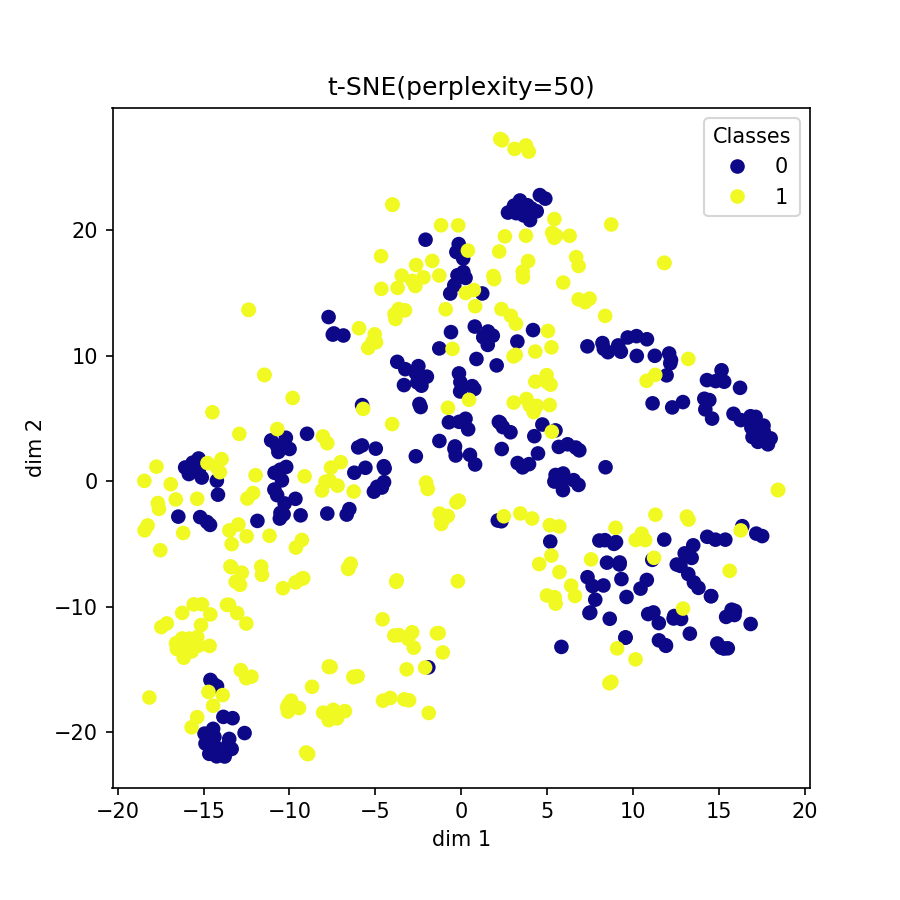 |
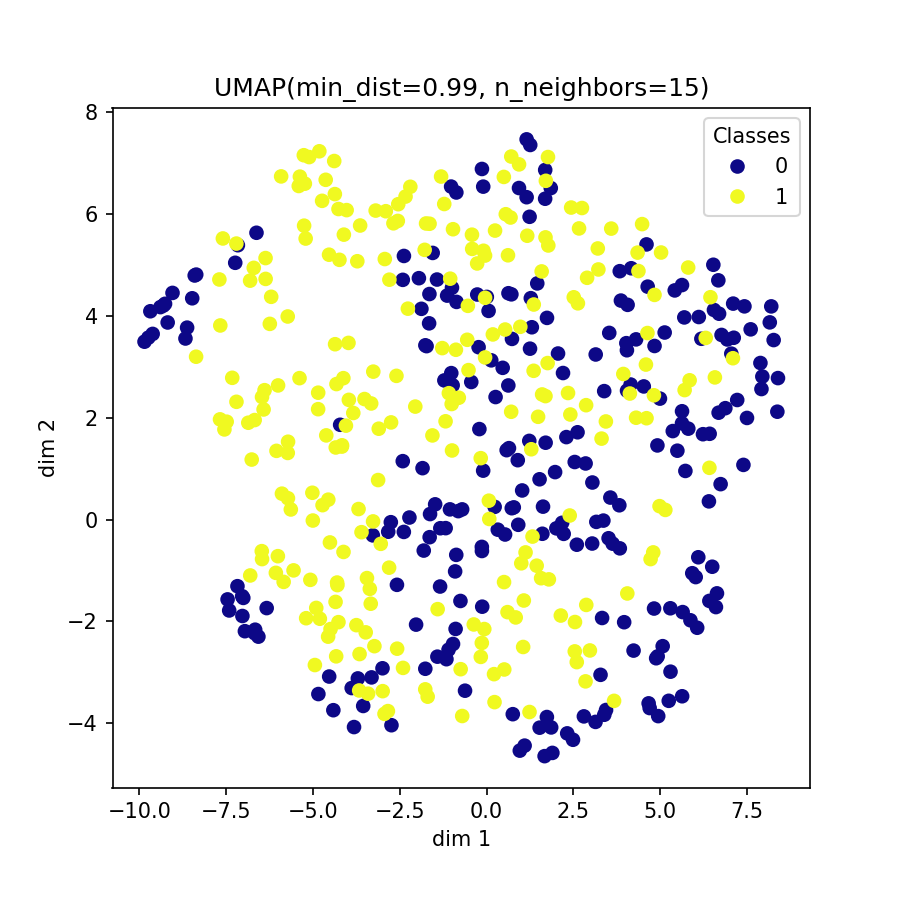 |
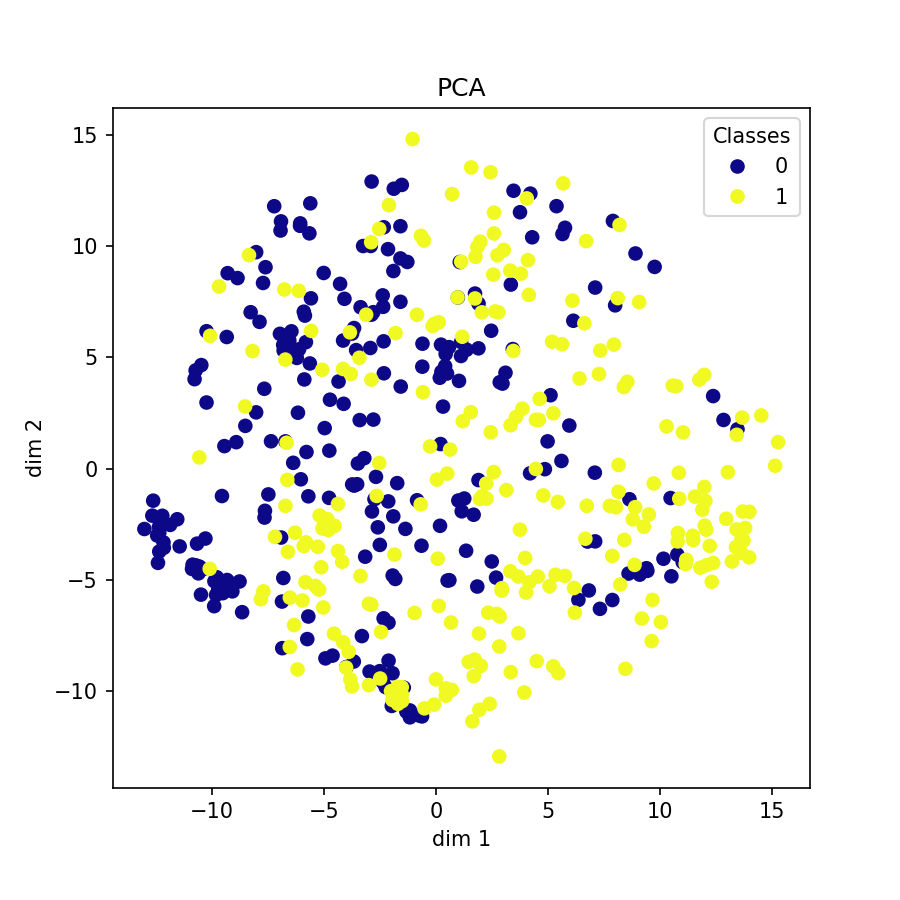 |
For more details about the project have a look at the README.md in the project directory /ECG-time-series.
Investigating which medical features from patient records (categorical, numerical and text) play an important role in the prediction of patient readmission. Comparing models using only numerical + categorical features, only text and both.
Results
| Cat./Num. Features |
|---|
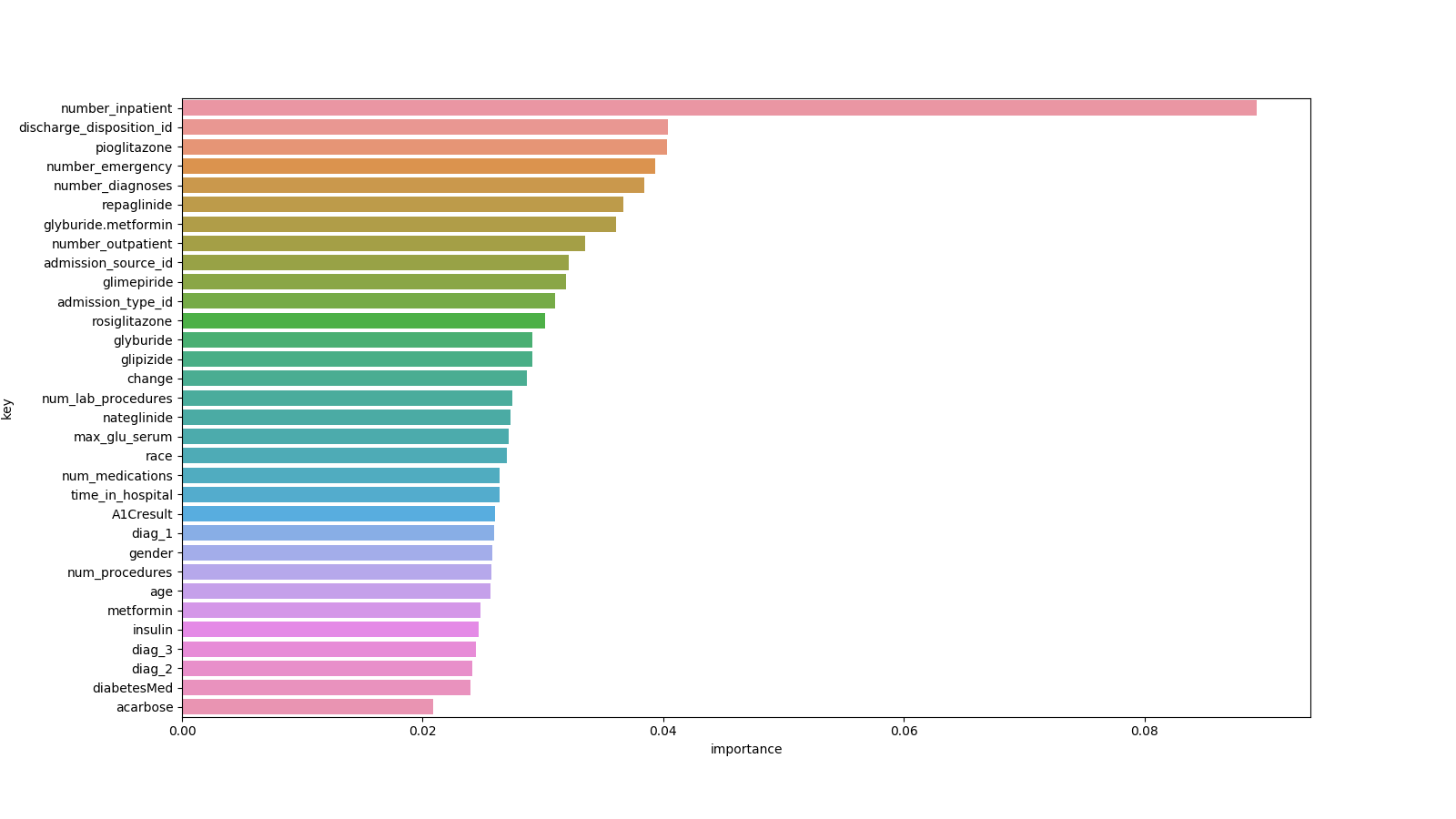 |
| Text Features |
|---|
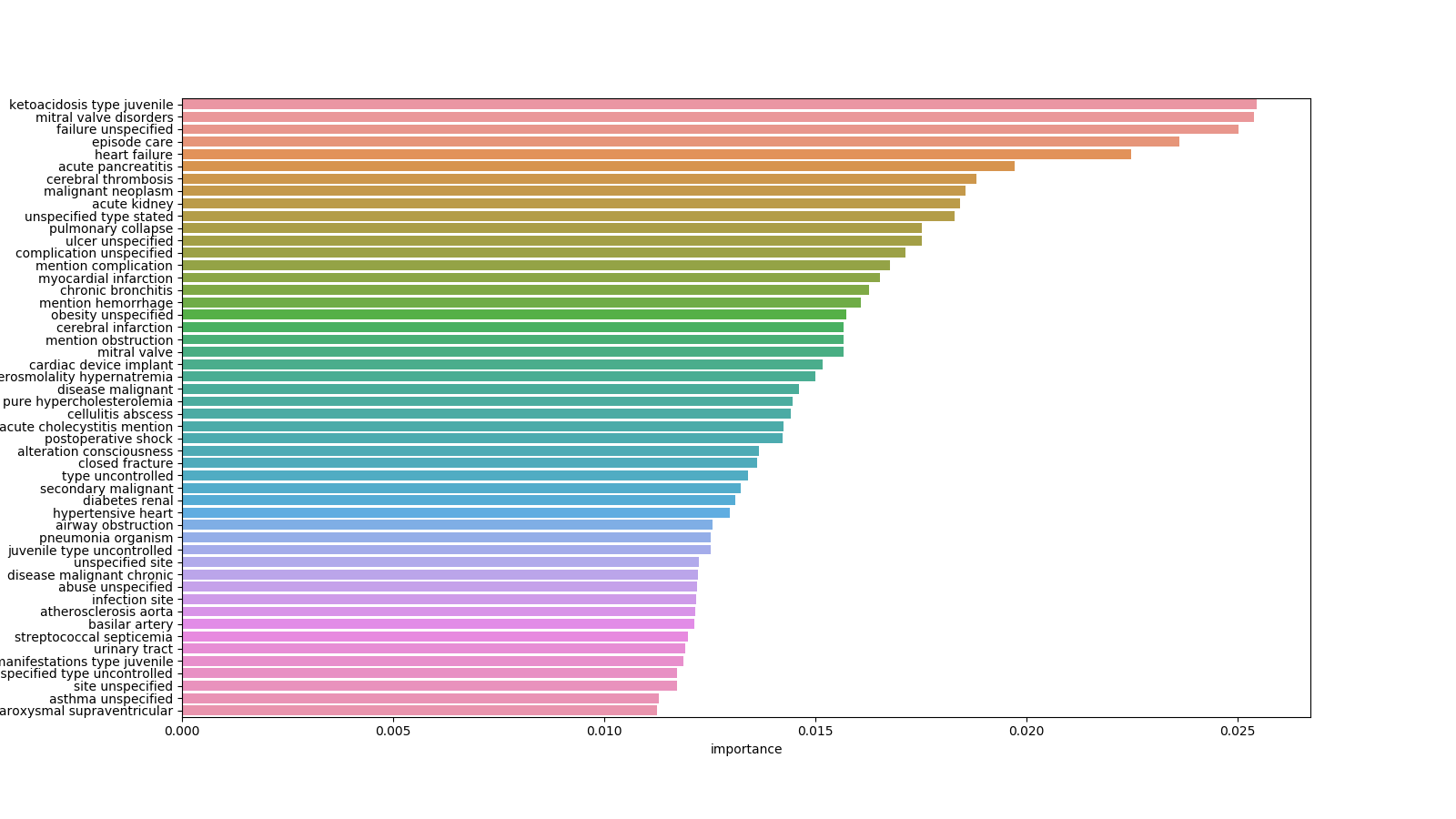 |
For more details about the project have a look at the README.md in the project directory /Diabetes-readmission.
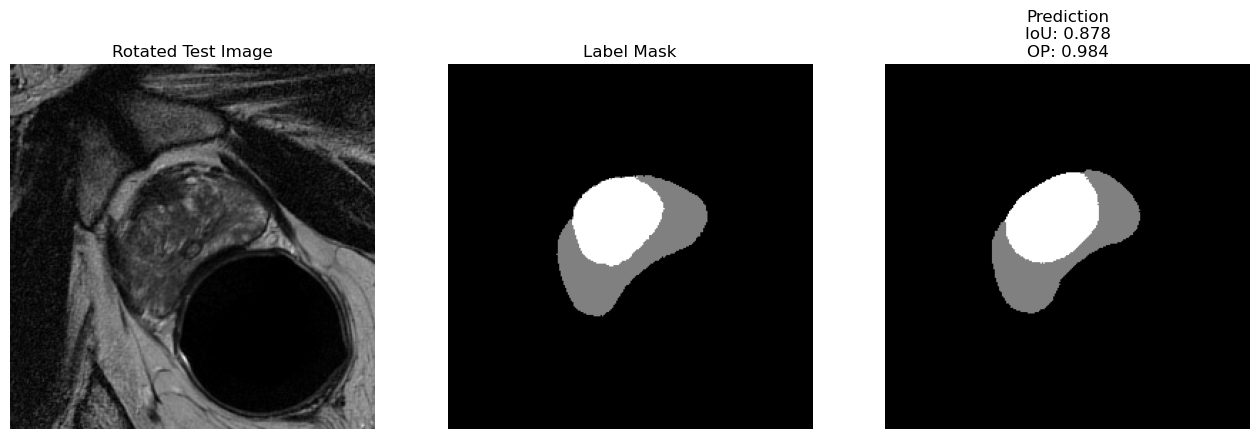
Using the U-Net neural network model [4] to segment MRI prostate images from the NCI-ISBI 2013 Challenge - Automated Segmentation of Prostate Structures into anatomical regions (Peripheral Zone & Central Gland). Part of the project was to perform hyperparameter-tuning and try different optimizers and loss-functions to reduce generalization error.
Results
| Example Test Image and Prediction |
|---|
For more details about the project have a look at the README.md in the project directory /Image-segmentation.
Splice site prediction is common problem in computational genome finding where it is desirable to find the splice sites that mark the boundaries of exons and introns in organisms whose cells have a nucleus enclosed within membranes (eukaryotes). This classification can then be used to predict a gene's structure, function, interaction or its role in a disease. Main challenge of this task was the high class imbalance of the splice sites.
Results
| C.Elegans DNA |
|---|
| Human DNA |
|---|
For more details about the project have a look at the README.md in the project directory /Splice-site-prediction.
pandas, numpy, scikit-learn, keras, matplotlib, xgboost, umap-learn, seaborn, keras-contrib, tensorflow-addons
[1] Mohammad Kachuee, Shayan Fazeli, and Majid Sarrafzadeh. "ECG Heartbeat Classification: A Deep Transferable Representation." arXiv preprint arXiv:1805.00794 (2018) .
[2] CVxTz's GitHub implementation: ECG_Heartbeat_Classification (link)
[3] Impact of HbA1c Measurement on Hospital Readmission Rates: Analysis of 70,000 Clinical Database Patient Records. https://doi.org/10.1155/2014/781670
[4] Ronneberger, Olaf & Fischer, Philipp & Brox, Thomas. (2015). U-Net: Convolutional Networks for Biomedical Image Segmentation. LNCS. 9351. 234-241. 10.1007/978-3-319-24574-4_28.
For this repository the cookiecuter data science project template is used slightly adapted to the our needs and requirements. Each of the four projects is in a separate folder which is a copy of the src directory.
├── LICENSE
├── Makefile <- Makefile with commands like `make data` or `make train`
├── README.md <- The top-level README for developers using this project.
├── data
│ ├── external <- Data from third party sources.
│ ├── interim <- Intermediate data that has been transformed.
│ ├── processed <- The final, canonical data sets for modeling.
│ └── raw <- The original, immutable data dump.
│
├── docs <- A default Sphinx project; see sphinx-doc.org for details
│
├── models <- Trained and serialized models, model predictions, or model summaries
│
├── notebooks <- Jupyter notebooks. Naming convention is a number (for ordering),
│ the creator's initials, and a short `-` delimited description, e.g.
│ `1.0-jqp-initial-data-exploration`.
│
├── references <- Data dictionaries, manuals, and all other explanatory materials.
│
├── reports <- Generated analysis as HTML, PDF, LaTeX, etc.
│ └── figures <- Generated graphics and figures to be used in reporting
│
├── requirements.txt <- The requirements file for reproducing the analysis environment, e.g.
│ generated with `pip freeze > requirements.txt`
│
├── setup.py <- makes project pip installable (pip install -e .) so src can be imported
├── src <- Source code for use in this project.
│ ├── __init__.py <- Makes src a Python module
│ │
│ ├── data <- Scripts to download or generate data
│ │ └── make_dataset.py
│ │
│ ├── features <- Scripts to turn raw data into features for modeling
│ │ └── build_features.py
│ │
│ ├── models <- Scripts to train models and then use trained models to make
│ │ │ predictions
│ │ ├── predict_model.py
│ │ └── train_model.py
│ │
│ └── visualization <- Scripts to create exploratory and results oriented visualizations
│ └── visualize.py
│
└── tox.ini <- tox file with settings for running tox; see tox.testrun.org
Project based on the cookiecutter data science project template. #cookiecutterdatascience