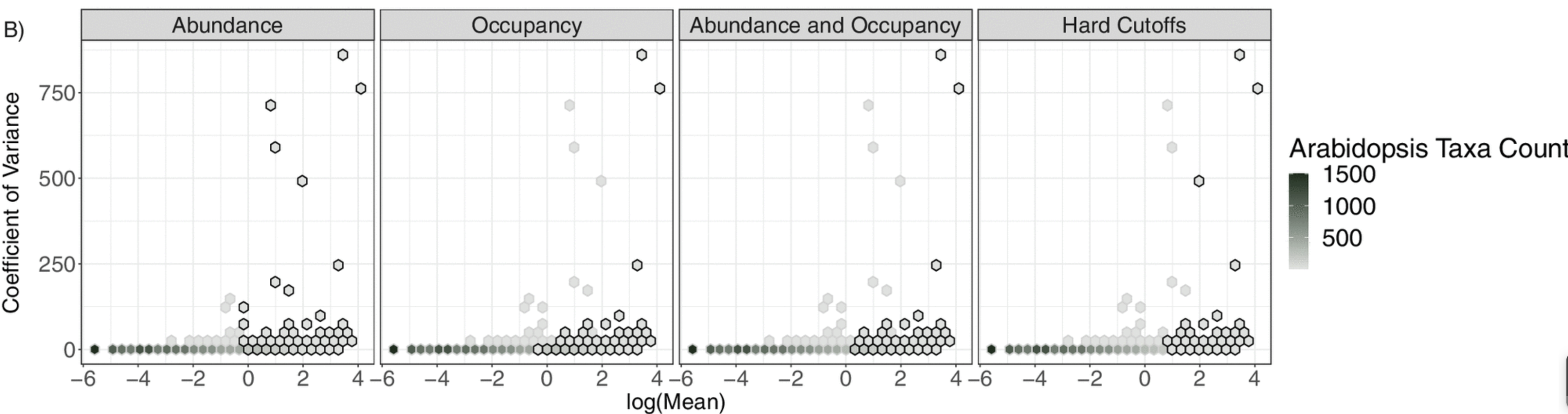
Companion package to manuscript: Comparative Analysis of Core Microbiome Assignments: Implications for Ecological Synthesis (DOI: https://doi.org/10.1128/msystems.01066-22). This package allows for the comparison of commonly implemented methods for core assignments in microbiome studies. Each of the four core assignment methods included can be customized to alter thresholds required for core inclusion. This allows researchers to alter cutoffs and determine if core assignment methods agree at certain cutoffs, facilitating comparison of multiple core assignment methods.
You can install CoreMicro from
# install.packages("devtools")
devtools::install_github("MayaGans/CoreMicro")
Using our built in arabidopsis dataset, you can create a dataframe describing taxa inclusion by method, and plot taxa inclusion using the function core_plots(). See our vignettes for further use cases.
library(CoreMicro)
arabidopsis %>% # use built in dataset
core_methods() %>% # create taxa presence absence table per core method
core_plots() # plot using hexbinsTo learn about how to use CoreMicro was used in Comparative Analysis of Core Microbiome Assignments: Implications for Ecological Synthesis (DOI: https://doi.org/10.1128/msystems.01066-22), check out the vignettes folder.
vignette("analysis_and_plots", "CoreMicro"): Learn how to use all functions exported by the package.