This repository contains Jupyter notebooks demonstrate how pydna can be used to plan, document and simulate virtually any cloning experiment.
The links below open a static version of each notebook as a web page directly on Github.
-
Construction of the YEp24PGK_XK vector using restriction and ligation.
-
Construction of the pGUP1 sequence using in-vivo homologous recombination
-
Assembly of the 32 Mumberg expression vectors
-
Assembly of the pGreenLantern-1 sequence.
-
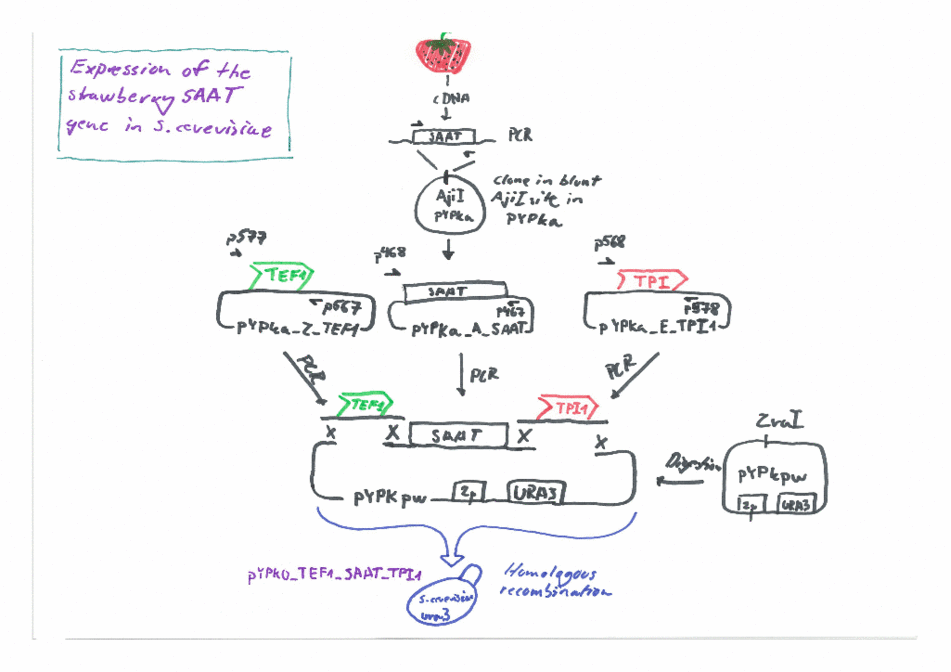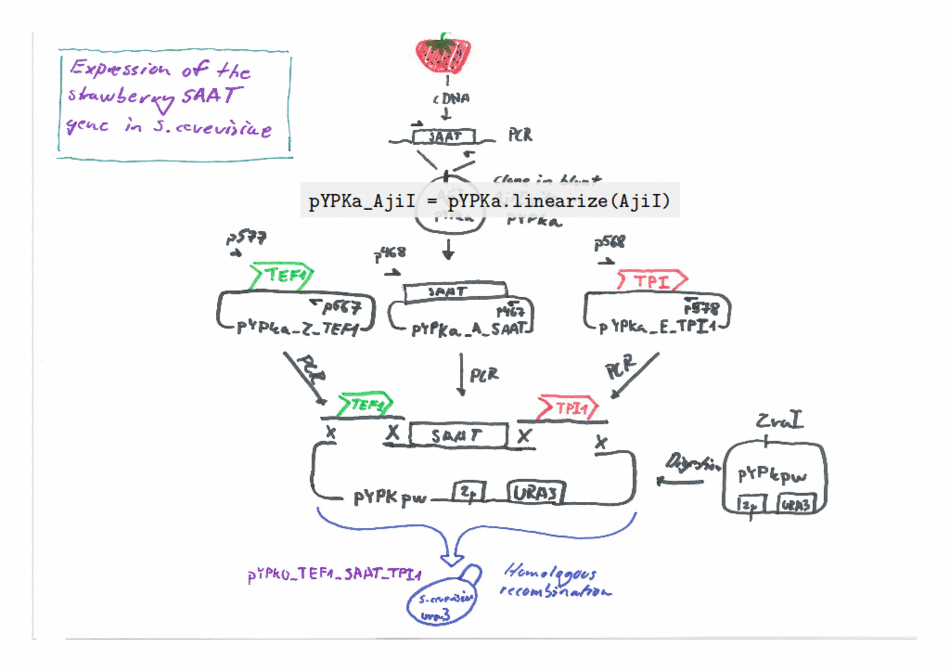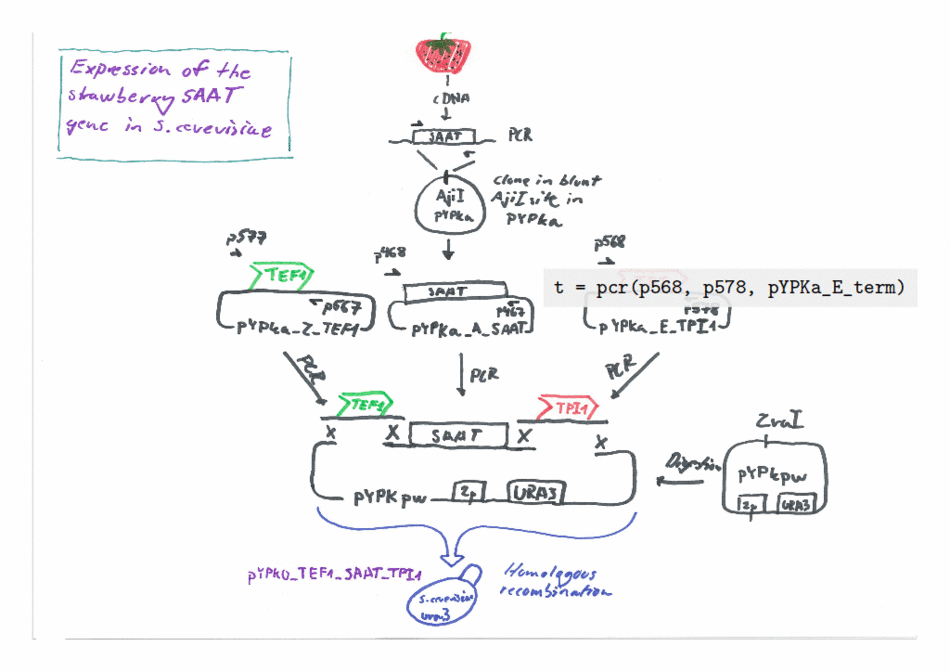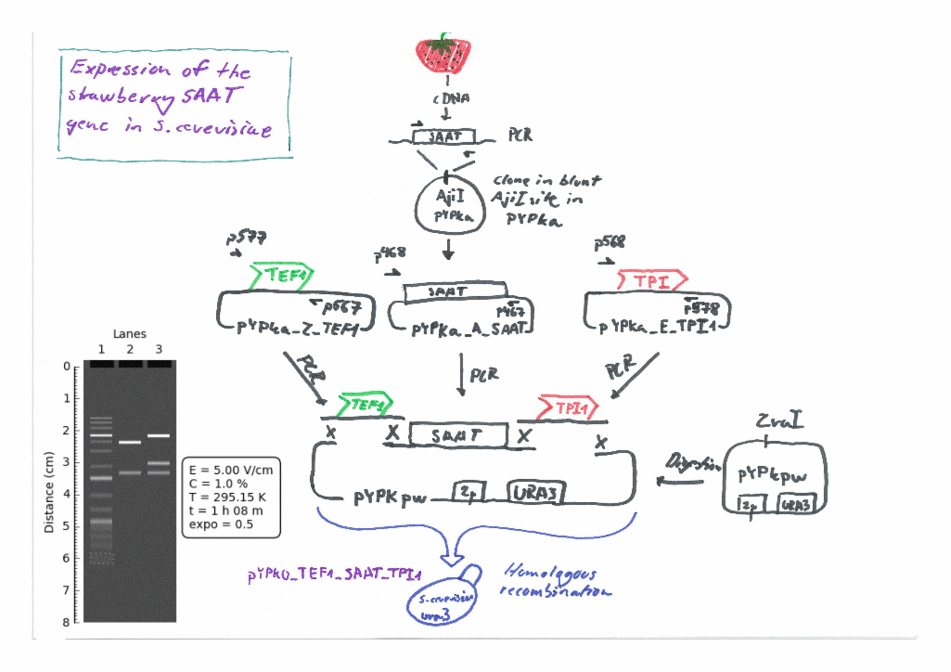
Expression of the Strawberry aat gene in Saccharomyces cerevisiae using the Yeast Pathway Kit
-
Gibson assembly example
-
Golden gate cloning example
Static versions of the notebooks can also be displayed as web pages through nbviewer.
Click on the red and black "launch binder" image above to run the notebooks on the cloud service mybinder. This way notebooks can be executed dynamically online without need to install anything on the users computer.
N.B. Mybinder runs on infrastructure provided for free. Please do not abuse the service. If you want to do extensive testing of these notebooks, please download this repository and run the notebooks locally (the notebooks will also run faster).
The Jupyter notebooks in this repository are tested using a continuous integration service. A fresh install of all depending software is performed once per month or whenever the files in this repository change. The tests compare the old, saved output from each code cell with a newly calculated output. If the content do not match for any cell, the badge above turns red. The bagde is also a link to the server where the tests are performed.
The contents of this repository is released under the MIT Licence
Use this Digital Object Identifier (DOI) when citing this repository.