ESTIpop is an R-wrapped C++ package to simulate and estimate the parameters of continuous-time Markov branching processes with time-dependent rates.
- GNU Scientific Library
- (OSX) brew install gsl with Homebrew or from (http://ftpmirror.gnu.org/gsl/).
- (Windows) download and extract the file local###.zip and create an environmental variable LIB_GSL to add the directory (see notes about Windows installation below for more details).
- (Linux) install libgsl0-dev and gsl-bin.
- Rtools (Windows only)
- devtools
Rtools contains the necessary resources to compile C++ files when installing packages in R. GSL is also required which can be downloaded from (http://www.stats.ox.ac.uk/pub/Rtools/goodies/multilib/local323.zip). After downloading, unzip to your R directory. Depending on if your computer is 32 or 64-bit, move the library files from local###/lib/i386 (32-bit) or local###/lib/x64 (64-bit) to local###/lib.
To set the environmental variable LIB_GSL on a Windows 7/10 computer, go to “Advanced system settings” in Control Panel > System and Security > System and click Environmental Variables. Create a new system variable with
- Variable Name: LIB_GSL
- Variable Value: “C:/path/to/local323” (include quotes)
- ggplot2
- R.utils
To install in R, type:
install.packages("devtools")
devtools::install_git("https://github.com/Michorlab/estipop.git")ESTIpop is an R package that estimates the parameters of continuous-time Markov branching processes (CTMBPs) with constant or time-dependent rates. A CTMBP is a stochastic process in which individuals live for a random amount of time before generating some number of offspring, each of which continues the process. Individuals can belong to one of a finite number of types. Each type has its own continuous lifetime distribution, as well a distirbution that governs the number and types of offspring it produces. The process is Markov because an individual’s remaining lifetime is independent of the individual’s age. However, the rates at which individuals die and reproduce can still vary with time, analogous to the nature of arrivals in a time-inhomogenous Poisson process.
ESTIpop also provides methods to simulate continuous-time Markov branching processes. Simulated processes can have arbitrary numbers of types and transitions, and may have rates which are constant or time-dependent.
ESTIpop uses a common representation of branching process models for
simluation and estimation. A branching process model is represented by a
process_model object containing one or more transition objects, and
each transition object is associated with a rate object.
A rate object represents the rate at which some event occurs, and may
be a function of time and multiple other parameters. The constructor
rate(exp) takes an R expression which describes how the rate depends
on time and other parameters. For example:
r1 = rate(3.5) # a constant, known rate
r2 = rate(2*t) # a rate which increases linearly with time
r3 = rate(1.5*params[1]) #a rate which depends on one unknown parameter
r4 = rate(params[1] + params[2]*exp(-.5*t)) # a rate which depends on 2 unknown
# parameters and timeIt is important to note that the variable t and the params vector
are not variables which we have defined previously in the program. The
rate constructor acts on an expression which encodes how the rate
evolves with time and various other parameters. We will be able to
evaluate this expression later when we plug in parameter values at a
specific time.
The rate constructor expects expressions which conform to the
following subset of the R grammar, and will throw an error at other
inputs.
| Type | Allowed Symbols |
|---|---|
| Unary Operators | + - () |
| Binary Operators | + - / * > >= < <= |
| Functions | sin(), cos(), log(), exp() |
| names | params[i], t |
| numerics | any scalar numeric value |
A transition object represents a type of birth or death event which
can occur in the branching process model. A transition object has 3
attibutes:
parent: The type of the parent which dies and is removed from the population when the transition occursoffspring: The number of each type of offspring which are produced in the transition. This should be and-length vector wheredis the number of types in the population.rate: Arateobject (described above) governing how quickly this transition occurs
A transition object can be constructed as follows:
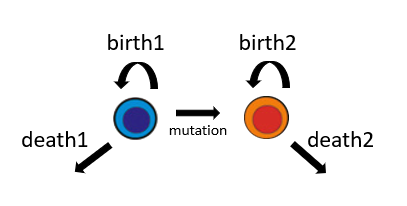
r = rate(.5*params[1] + params[2]*exp(-t))
#a type 2 parent dies and produces 2 offspring of different types
trans = transition(rate = r, parent = 1, offspring = (2,0)) This transition is illustrated in the following figure:
A process_model object is a collection of transition objects which
together make up a branching process system. A process_model object is
constructed from a sequence of transition objects which can occur in
the system. All of the transition objects must assume the same number
of types, and the process_model constructor will throw an error if
this is not the
case.
#simple one-type birth death model with rates that decay exponentially over time
birth = rate(params[1]*exp(-params[3]*t))
death = rate(params[2]*exp(-params[3]*t))
model = process_model(transition(rate = birth, parent = 1, offspring = 2),
transition(rate = death, parent = 1, offspring = 0))Note that model still retains all of the time- and
parameter-dependence of its constituent transitions. Also notice that,
since params[3] was used in both the birth and death rate, this model
has the constraint that birth and death rates decay at equal speed.
Armed with a process_model object, ESTIpop allows for the simulation
of this system under various parameters values. ESTIpop uses C++ for
high-performance simulation. In the case of time-dependent rates,
ESTIpop parses the expressions and translates them into dynamically
loaded C++ code.
The branch function is used to simulate the system specified by a
process_model object. It’s parameters are as
follows:
| Argument | Type | Description | Optional? |
|---|---|---|---|
model |
process_model object |
The system to simulate | No |
params |
numeric vector | The parameters to plug into the system during simulation | No |
init_pop |
numeric vector | The population at the start of the simulation (time 0) | No |
time_obs |
numeric vector | The timepoints at which to record the state of the population | No |
reps |
numeric scalar | The number of times to run the simulation | No |
silent |
logical | Whether to silence intermediate printouts form the C++ simulator | Yes |
keep |
logical | Whether to keep logs files from the C++ simulation | Yes |
seed |
logical | A seed for the random number generator | Yes |
The following examples demonstrate ESTIPop’s simulation features:
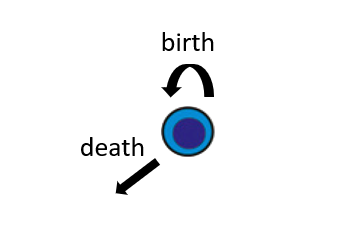
We’ll start by simulation the one-type birth-death model shown in the following figure. In this model, a population of a single type experiences birth events, in which an individual from the population is chosen to replicate, and death events, in which an individual from the population is chosen for removal. To test our estimation procedure, we begin by simulating data using functions available in ESTIpop. We initiate the population with size 100 and allow it expand for 5 units of time with birth parameter 1 and death parameter 0.7. Using the following code, we generate 1,000 samples from this process.
library(estipop)
#Create a one-type birth-death model where the birth rate increases over time
model = process_model(transition(rate = rate(params[1] - params[2]*exp(-params[3]*t)),
parent = 1, offspring = 2),
transition(rate = rate(params[4]), parent = 1, offspring = 0))
#Specify the time points at which we will record the state of the simulation
time = seq(0,20,.5)
#The initial size of the simulated population
init_pop = c(500)
#Parameters to plug into the model for this simulation run
params = c(.3,.25,.1, .2)
#Numer of times to run the simulation
reps <- 1000
res = branch(model = model, params = params, init_pop = init_pop,
time_obs = time, reps = reps)
#Plot a single replication of the simulation
single <- dplyr::filter(res, rep == 1)
plot(single$time, single$type1, xlab = "time", ylab = "population size")res now contains a dataframe with the result of the simulation. We can
now analyze the results of the simulation.
res <- dplyr::filter(res, time == 20) #just take the last timepoint
#compute the theoretical moments of the population distribution
mom <- compute_mu_sigma(model, params, 0, 20, init_pop)
print(paste("Sample Mean: ", mean(res$type1)))## [1] "Sample Mean: 426.535"
print(paste("True Mean: ", mom$mu))## [1] "True Mean: 425.364259883693"
print(paste("Sample Variance: ", var(res$type1)))## [1] "Sample Variance: 4502.39116616617"
print(paste("True Variance: ", mom$Sigma))## [1] "True Variance: 4386.38004944714"
We now consider the two-type process shown in the following figure. Here, each of the two population types has separate birth and death rates, as well a mutation rate from the type 1 population to the type 2 population. We’ll again simulate 1000 examples and compare the results with the theoretical moments.
library(estipop)
time = 5
init_pop= c(100, 0)
params = c(.4,.1,.7,.1, .3)
#time-homogenous two-type model
model = process_model(transition(rate = rate(params[1]), parent = 1, offspring = c(2,0)),
transition(rate = rate(params[2]), parent = 1, offspring = c(0,0)),
transition(rate = rate(params[3]), parent = 2, offspring = c(0,2)),
transition(rate = rate(params[4]), parent = 2, offspring = c(0,0)),
transition(rate = rate(params[5]), parent = 1, offspring = c(1,1)))
res = branch(model, params, init_pop, time, 1000)Now we analyze the simulation results and compare them with theory
mom = compute_mu_sigma(model, params, 0, 5, init_pop)
print(paste("Sample Mean: ", toString(c(mean(res$type1), mean(res$type2)))))## [1] "Sample Mean: 448.151, 1571.425"
print(paste("True Mean: ", toString(mom$mu)))## [1] "True Mean: 448.168943658686, 1560.38542784701"
print(paste("Sample Variance: ", toString(c(var(res$type1), var(res$type2)))))## [1] "Sample Variance: 2756.96316216216, 41241.8542292292"
print(paste("Sample Covariance:", cov(res$type1, res$type2)))## [1] "Sample Covariance: 4801.0769019019"
print(paste("True Variance: ", toString(diag(mom$Sigma))))## [1] "True Variance: 2600.64200784677, 39457.2214316809"
print(paste("True Covariance: ", toString(mom$Sigma[1,2])))## [1] "True Covariance: 4527.31533883025"
Given process_model object and a set of datapoints generated from that
model, ESTIpop can estimate the model’s unknown parameter values.
The estimate and estimate_td functions are used to estimate the
parameters of process_model object. The arguments these function as as
follows:
| Argument | Type | Description |
|---|---|---|
model |
process_model object |
The system whose parameters are being estimated |
init_pop |
N x d matrix |
The initial population in each of the observations in the dataset |
final_pop |
N x d matrix |
The final observed population in each observation |
start_times |
N x 1 vector |
The time at which each entry in init_pop was observed |
end_times |
N x 1 vector |
The time at which each entry in final_pop was observed |
initial_ params |
m x 1 vector |
The parameter values at which to initialize the estimation procedure |
lower |
m x 1 vector |
Lower bounds on each parameter to be estimated |
upper |
m x 1 vector |
Uppose bounds on each parameter to be estimated |
trace |
logical | Whether to enable output during optimization |
The estimate function is a wrapper for the estimate_td function.
estimate only requires the elapsed time between the initial and final
obervations of each entry in the dataset, rather then the exact times of
each observation. estimate can be used when the rates of a model do
not depend on time.
The following examples demonstrate ESTIPop’s estimation features:
We now return to the one-type time-dependent model explored in the simulation section. In this model the birth rate declines as an unknown exponential function while the death rate remains constant. This could represent the dynamics of a population recovering from a drug for which we want to estimate a pharmacokinetic model. First we will simulate the model, this time with far fewer replications:
library(estipop)
#Create a one-type birth-death model where the birth rate increases over time
model = process_model(transition(rate = rate(params[1] - params[2]*exp(-params[3]*t)),
parent = 1, offspring = 2),
transition(rate = rate(params[4]), parent = 1, offspring = 0))
#Specify the time points at which we will record the state of the simulation
time = seq(0,20,1)
#The initial size of the simulated population
init_pop = c(500)
#Parameters to plug into the model for this simulation run
params = c(.3,.25,.1, .2)
#Numer of times to run the simulation
reps <- 20
res = branch(model = model, params = params, init_pop = init_pop,
time_obs = time, reps = reps)
simdat <- format_sim_data(res, ntypes = 1)The last line converts the dataset into the format expected by our estimation functions. Note that, because the process is Markov, we can compute the likelihood of the entire dataset by computing the likelihood of each observation conditioned on the previous observation. We now estimate the parameters of the model:
est = estimate_td(model, init_pop = simdat$type1_prev,
final_pop =simdat$type1,
start_times = simdat$prev_time,
end_times = simdat$time,
initial = runif(4,0,.5),
lower = rep(0,4),
upper = rep(.5,4),
method = "L-BFGS-B")print(est$par)## [1] 0.2632411 0.2414000 0.1098404 0.1774179
After estimation we recover parameter values very close to ground truth.
We’ll now estimate the parameters of the two-type birth-death-mutation model we simulated earlier.
time = seq(0,10,1)
initial = c(100, 0)
params = c(.4,.1,.7,.1, .3)
model = process_model(transition(rate = rate(params[1]), parent = 1, offspring = c(2,0)),
transition(rate = rate(params[2]), parent = 1, offspring = c(0,0)),
transition(rate = rate(params[3]), parent = 2, offspring = c(0,2)),
transition(rate = rate(params[4]), parent = 2, offspring = c(0,0)),
transition(rate = rate(params[5]), parent = 1, offspring = c(1,1)))
res = branch(model, params, initial, time, 30)
simdata = format_sim_data(res, model$ntypes)
est = estimate(model, init_pop = cbind(simdata$type1_prev, simdata$type2_prev),
final_pop = cbind(simdata$type1, simdata$type2),
times = simdata$dtime,
initial = runif(5,0,.5),
lower = rep(1e-5,5),
upper = rep(1,5),
method = "L-BFGS-B")print(est$par)## [1] 0.4084356 0.1097101 0.6591385 0.0601875 0.3050673
Once again we estimate values close to ground truth.
The two-type drug resistance model is a four-type process in which two types of mutation are possible. The type 0 population harbors no mutations, the type 1 population harbors mutation 1, the type 2 population harbors mutation 2, and the type 12 population harbors both mutations 1 and 2. Each type undergoes birth and death events, as well as the possibility for further mutation as illustrated in the following figure.
We now simulate and estimate this model. To reduce the number of parameters, we assume that all types share the same death rate.
time = seq(0,10,1)
initial = c(100, 0,0,0)
params = c(.4,.7,.5,.2,.3,.1,.4,.3, .3)
model = process_model(
transition(rate = rate(params[1]), parent = 1, offspring = c(2,0,0,0)),
transition(rate = rate(params[2]), parent = 2, offspring = c(0,2,0,0)),
transition(rate = rate(params[3]), parent = 3, offspring = c(0,0,2,0)),
transition(rate = rate(params[4]), parent = 4, offspring = c(0,0,0,2)),
transition(rate = rate(params[5]), parent = 1, offspring = c(1,1,0,0)),
transition(rate = rate(params[6]), parent = 1, offspring = c(1,0,1,0)),
transition(rate = rate(params[7]), parent = 2, offspring = c(0,1,0,1)),
transition(rate = rate(params[8]), parent = 3, offspring = c(0,0,1,1)),
transition(rate = rate(params[9]), parent = 1, offspring = c(0,0,0,0)),
transition(rate = rate(params[9]), parent = 2, offspring = c(0,0,0,0)),
transition(rate = rate(params[9]), parent = 3, offspring = c(0,0,0,0)),
transition(rate = rate(params[9]), parent = 4, offspring = c(0,0,0,0)))
res = branch(model, params, initial, time, 30)
simdata = format_sim_data(res, model$ntypes)
init_pop = cbind(simdata$type1_prev, simdata$type2_prev,
simdata$type3_prev, simdata$type4_prev)
final_pop = cbind(simdata$type1, simdata$type2,
simdata$type3, simdata$type4)
est = estimate(model, init_pop = init_pop,
final_pop = final_pop,
times = simdata$dtime,
initial = rbeta(9,2,1),
lower = rep(1e-5,9),
upper = rep(1,9),
method = "L-BFGS-B")print(est$par)## [1] 0.3882471 0.6848637 0.4824548 0.1992096 0.2986555 0.1032464 0.3927194 0.3187989 0.2878569
Once again, our estimates agree closely with ground truth.
/michor/projects/estipop-jeremy/estipop/README_files/figure-gfm/sup1-1.png)