The code for the paper: PharmKG -- A Dedicated Knowledge Graph Benchmark for Biomedical Data Mining
The code was partly built based on Pykeen and KG-reeval. Thanks a lot for their code sharing!
The initial development was made by Aladdin Healthcare Technologies Ltd., Sun Yat-sen University and Mind Rank AI. All Rights Reserved.
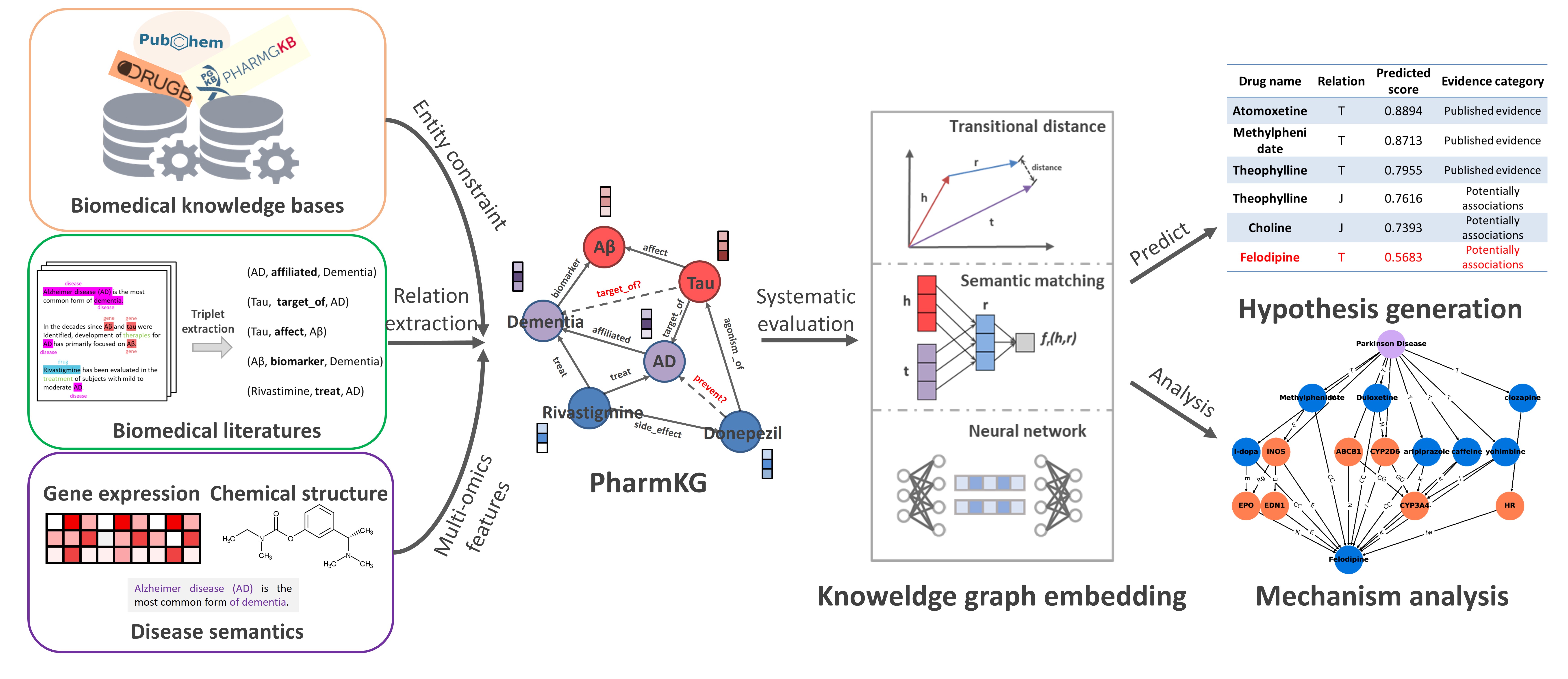
PharmKG is a multi-relational, attributed biomedical knowledge graph, comsed of more than 500 thousands individual interconnectons between genes, drugs and diseases, with 29 relation types over a vocabulary of ~8000 disambiguated entites.
Raw dataset of PharmKG was hosted on zenodo. And in the experiments we used the cleaned PharmKG-8K dataset. The detailed information can be found in PharmKG_original.zip
| Dataset | Train | Test | Valid | Entities | Triplets |
|---|---|---|---|---|---|
| PharmKG-8k | 400788 | 49536 | 50036 | 7601 | 500958 |
| PharmKG-Raw | - | - | - | 188296 | 1093236 |
| Type | DrugBank | TTD | OMIM | PharmGKB | GNBR | PharmKG |
|---|---|---|---|---|---|---|
| Chemical | 1208 | 1347 | - | 615 | 1442 | 1497 |
| Disease | - | 399 | 987 | 419 | 1001 | 1346 |
| Gene | 1166 | 741 | 2320 | 1674 | 4716 | 4758 |
| Category | Model | Hits@N | |||||
|---|---|---|---|---|---|---|---|
| MRR | N=1 | N=3 | N=10 | N=100 | |||
| Distance-Based | TransE | 0.091 | 0.034 | 0.092 | 0.198 | 0.524 | |
| TransR | 0.075 | 0.030 | 0.071 | 0.155 | 0.510 | ||
| Semantic Matching | RESCAL | 0.064 | 0.023 | 0.057 | 0.122 | 0.413 | |
| ComplEx | 0.107 | 0.046 | 0.110 | 0.225 | 0.552 | ||
| Distmult | 0.063 | 0.024 | 0.058 | 0.133 | 0.461 | ||
| Neural Network | ConvE | 0.086 | 0.038 | 0.087 | 0.169 | 0.425 | |
| ConvKB | 0.106 | 0.052 | 0.107 | 0.209 | 0.548 | ||
| RGCN | 0.067 | 0.027 | 0.062 | 0.139 | 0.236 | ||
| Proposed | HRGAT –w/o | 0.138 | 0.068 | 0.148 | 0.275 | 0.586 | |
| HRGAT | 0.154 | 0.075 | 0.172 | 0.315 | 0.649 | ||
Under the "PharmKG-D/model/pykeen/pykeen" directory, type python setup.py install --user to compile the pykeen package.
Run code python PharmKG-D/data/preprocess.py
The data for the preprocess.py can be found here
The embedding data can be found here
TransE, TransR, DistMult, ComplEx, RESCAL
Run code python PharmKG-D/model/pykeen/train.py --model <model_name> --save_path <path> under the root directory of this repository. <model_name> is name of the model you are going to train. <path> is the path to a json file containing the output results.
ConvE, ConvKB, HRGAT
Model training can be started by running the following scripts:
ConvE:
sh PharmKG-D/model/ConvE/run.shConvKB:
sh PharmKG-D/model/ConvKB/run.shHRGAT:
sh PharmKG-D/model/HRGAT/run.sh