DrugTax: package for drug taxonomy identification and explainable feature extraction
A.J.Preto, Paulo C. Correia and Irina S. Moreira
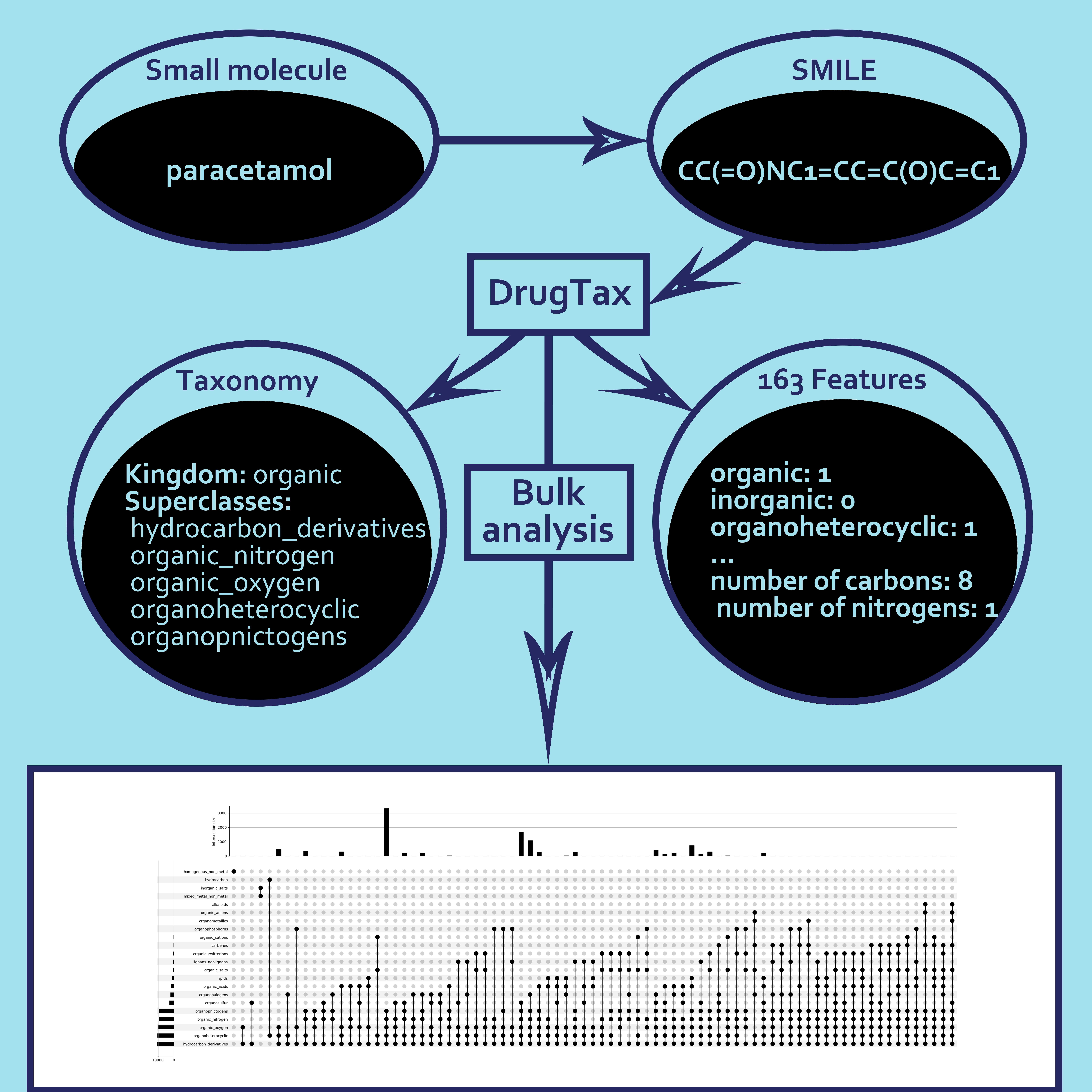
Abstract: DrugTax is an easy-to-use Python package for small molecule detailed characterization. It extends a previously explored chemical taxonomy making it ready-to-use in any Artificial Intelligence approach. DrugTax leverages small molecule representations as input in one of their most accessible and simple forms (SMILEs) and allows the simultaneously extraction of taxonomy information and key features for big data algorithm deployment. In addition, it delivers a set of tools for bulk analysis and visualization that can also be used for chemical space representation and molecule similarity assessment. DrugTax is a valuable tool for chemoinformatic processing and can be easily integrated in drug discovery pipelines. DrugTax can be effortlessly installed via PyPI (https://pypi.org/project/DrugTax/) or GitHub (https://github.com/MoreiraLAB/DrugTax).
To install DrugTax, first make sure you have python 3.6.x, or above, installed.
pip install drugtax upsetplot==0.6.0 pandas==1.1.5 matplotlib==3.3.4 pubchempy==1.0.4
In order to ensure all necessary packages are installed.
Firstly, import DrugTax
import drugtax
The basic usage of DrugTax stems from the DrugTax class, which takes as input a single SMILE.
molecule = drugtax.DrugTax("OC(=O)C1=C(C(O)=O)C(C(O)=O)=C(C(O)=O)C(C(O)=O)=C1C(O)=O")
The molecule object now has a series of useful properties such as:
molecule.smile: allows the user to check the SMILE at any timemolecule.superclasses: displays all the superclasses to which the input SMILE belongsmolecule.features: retrieves simple and explainable features that can be used on prediction tasks or dataset characterizationmolecule.kingdom: informs on whether the molecule is organic or inorganic
For superclass computation, instead of directly invoking the DrugTax class, it is possible to use retrieve_taxonomic_class on several different inputs. The example below shows an example using only a SMILEs list, however, it is also possible to feed a drugs list - in which case DrugTax leverages pubchempy to retrieve the isomeric SMILEs - or a file. This function outputs a table with the SMILEs and their respective taxonomy, as well as a summary table detailing how many of which superclass combinations are present on the dataset.
smiles_table, summary_table = drugtax.retrieve_taxonomic_class(["CCNO","CCC"], input_mode = "smiles_list", output_name = "testing", write_values = True)
The retrieve_taxonomic_class function has different arguments that can be used to pick input and output information for bulk analysis submission. These are:
input_data: this is the only mandatory argument, corresponding to either a smiles list, a drugs names list or a file.input_mode: depending on theinput_datathis argument needs to be changed. The default mode isfile, which requires an input.csvfile. This argument needs to be coupled withtarget_column, specifying the name of the column containing the input SMILEs. To input a list of SMILE,input_modeneeds to be changed tosmiles_list. To input a list of drug names from which isomeric SMILEs are to be retrieved with the aid ofpubchempy, the user needs to changeinput_modetodrugs_list.target_column: this argument needs to be specified when using thefileinput mode.output_name: if the user wishes to save the file, he should specify this argument.write_values: with defaultFalse, the user needs to change this argument toTrueif he wishes to save the output tables.input_sep: if the input mode isfile, the user can change this argument depending on the column separator on the input file.
In order to visualize the data retrieved from bulk analysis, DrugTax leverages UpSetPlot, a package designed to allow the visualization of a large number of intersecting sets. This computation requires a file generated in the above Bulk Analysis sections. When writing the files, the summary table will be the one with the termination *_assess.csv, the beginning of the name depends on the users chosen output_name. This file is the one that can be fed to the plot_categories function.
drugtax.plot_categories("testing_assess.csv", output_name = "plot")
The plot_categories function has three arguments:
input_file: the name of the*_assess.csvpreviously retrieved.output_name: a name for the output*.pngfile.threshold: with default 1, this argument triggers an aggregation of low populated superclass combinations to their above counterpart.thresholdis the minimum number of entries in the file for it to be aggregated.element_size: with default 100, this argument defines the size of labels and titles on the plot.
If you use this work, please cite:
A.J.Preto, Paulo C. Correia & Irina S. Moreira, DrugTax: package for drug taxonomy identification and explainable feature extraction, Journal of Cheminformatics, 14(73): 2022. https://jcheminf.biomedcentral.com/articles/10.1186/s13321-022-00649-w