Pfam seed random split protein sequence multi-class classification using deep learning visit blog on medium link - https://medium.com/@jangidajay271/pfam-seed-random-split-protein-sequence-multi-class-classification-using-deep-learning-a0b172015c67
Understanding the relationship between amino acid sequence and protein function is a long-standing problem in molecular biology with far-reaching scientific implications. Despite six decades of progress, state-of-the-art techniques cannot annotate 1/3 of microbial protein sequences, hampering our ability to exploit sequences collected from diverse organisms. To address this, we report a deep learning model that learns the relationship between unaligned amino acid sequences and their functional classification across all 17929 families of the Pfam database. Our model co-locates sequences from unseen families in embedding space, allowing sequences from novel families to be accurately annotated. These results suggest deep learning models will be a core component of future protein function prediction tools. Predicting the function of a protein from its raw amino acid sequence is the critical step for understanding the relationship between genotype and phenotype. As the cost of DNA sequencing drops and metagenomic sequencing projects flourish, fast and efficient tools that annotate open reading frames with the function will play a central role in exploiting this data.
you can find the flask css code for this in repo link here - https://github.com/Mrjaggu/flask-protein-sequence
Classifying given protein sequence of amino acid to one of the family accession. This model can be used for prediction of a given protein sequence of amino acid. The model will generate a number of classes probability values corresponding to the number of class or family accession. The highest probability value to the corresponding class will be the predicted class for that protein sequence of amino acid.
Objective:
Our objective is to predict the given protein sequence of amino acid as accurate as possible.
Constraints:
Interpretability: Interpretability is important for given protein sequence of amino acid it should predict correctly.
Latency: Given protein sequence of amino acid it should predict correct class so there is no need for high latency.
Accuracy: Our goal is to predict the given protein sequence of amino acid as accurate as possible. Higher the test accuracy, the better our model will perform in the real world.
This is a multi-class classification problem with 17929 different classes, so we have considered two performance metrics: Multi-Class Log-loss: We have used deep learning model with the cross-entropy layer in the end with 17929 softmax units, so, therefore, our goal is to reduce the multi-class log loss/cross-entropy loss. Accuracy: This tells us how accurately our model performs in predicting the expressions.
The data is provided by kaggle so we can directly download from given link Kaggle link — https://www.kaggle.com/googleai/pfam-seed-random-split
The approach used to partition the data into training/dev/testing folds is a random split.
Training data should be used to train your models.
Dev (development) data should be used in a close validation loop (maybe for hyperparameter tuning or model validation).
Test data should be reserved for much less frequent evaluations — this helps avoid overfitting on your test data, as it should only be used infrequently.
Each fold (train, dev, test) has a number of files in it. Each of those files contains CSV on each line, which has the following fields:
sequence: HWLQMRDSMNTYNNMVNRCFATCIRSFQEKKVNAEEMDCTKRCVTKFVGYSQRVALRFAE
family_accession: PF02953.15
sequence_name: C5K6N5_PERM5/28–87
aligned_sequence: ….HWLQMRDSMNTYNNMVNRCFATCI………..RS.F….QEKKVNAEE…..MDCT….KRCVTKFVGYSQRVALRFAE
family_id: zf-Tim10_DDP
sequence: These are usually the input features to your model. Amino acid sequence for this domain. There are 20 very common amino acids (frequency > 1,000,000), and 4 amino acids that are quite uncommon: X, U, B, O, Z.
family_accession: These are usually the labels for your model. Accession number in form PFxxxxx.y (Pfam), where xxxxx is the family accession, and y is the version number. Some values of y are greater than ten, and so ‘y’ has two digits.
family_id: One-word name for the family.
sequence_name: Sequence name, in the form “$uniprot_accession_id/$start_index-$end_index”.
aligned_sequence: Contains a single sequence from the multiple sequence alignment (with the rest of the members of the family in seed, with gaps retained.
Generally, the family_accession the field is the label, and the sequence (or aligned sequence) is the training feature.
This sequence corresponds to a domain, not a full protein.
The contents of these fields are the same as to the data provided in Stockholm format by PFam atftp://ftp.ebi.ac.uk/pub/databases/Pfam/releases/Pfam32.0/Pfam-A.seed.gz
We have used almost all the library of deep learning that we used normally like pandas, numpy, sklearn, Keras, etc. We can install by simply using
pip install ‘specify package name here’
We have done some pre-processing like lowercase the sequence and have used max sequence length of 100 and converting text data into numerical form by using sklearn countvectorizer or else our defined function of creating a dictionary and mapping char to index form and index to char form.
So finally we have reached to our last process of model fit. So hereby we have completed 90% of the process. So our main objective here is to give a protein sequence one by one and reduce cross-entropy loss. We have designed our own neural network architecture as follow:
As we can see we have input shape (100,24) i.e (None,100,24) since we have max sequence length of 100 for every protein sequence of amino acid and it’s one hot encoded so it’s like for example: for eg: we have shape (1,2,3) so one hot encode of this look like [[1 0 0],[0 1 0],[0 0 1]]
Here 1 describe where the char index is presented. So in this way, our all train, test and dev data are encoded and represented.
From the architecture, we can observe that we have 4 convolution layer of 1D, Dense layer, Maxpooling layer, Dropout layer, BatchNormalization layer, Activation and Flatten layer. We have specified weight initialization. If you do not specify the weight initialization method explicitly in Keras, it uses Xavier initialization also known as Glorot initialization. The aim of all these weight initializers is to find a good variance for the distribution from which the initial parameters are drawn.
About convolution layer,
X = Conv1D(32, 1 , strides=1,padding='valid', name='conv1d_1', kernel_initializer=glorot_uniform(seed=0))(input_s)
Here 32 specify 32 filters with a kernel size of 1 since we have used a various filter in convolution layer so that network learned better parameter. We have used padding=’ valid’ therefore there will be no padding.
We have used dropout layer between convolution layer since it helps to prevent overfitting. Here we have used different dropout rate i.e 0.5 defined that 0.5% of nodes will be dropped during the forward and backward pass since no parameter are learned during backpropagation. Since the network temporarily removes the units from the connections and removal of the number of nodes for connection is random. So we can get an optimal dropout rate by hyper-tuning or using various dropout rate. In our case, I observed during epochs we were overfitting so I tried with small to large dropout rate to avoid overfitting (0.1–0.5).
After that, we have a max-pooling layer where features are reduced in a meaningful way which helps in downsample of convolution layer and as we know the advantages like location invariance, scale invariance of a max-pool layer. In our network, we have used a 1D max pool layer of size 2 while iterating through a feature map. To understand more about the convolutional neural network and layers visit this link.
Finally, we have a dense layer which contains relu activation units. Since the output layer contains 2910 softmax units. It will generate 2910 class probability which is sum to 1. We will minimize the loss by feeding it back during backpropagation. So in this way, our neural network model will get trained to classify the protein sequences.
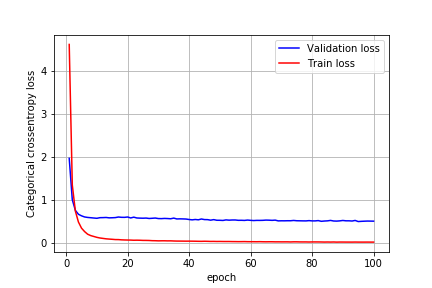
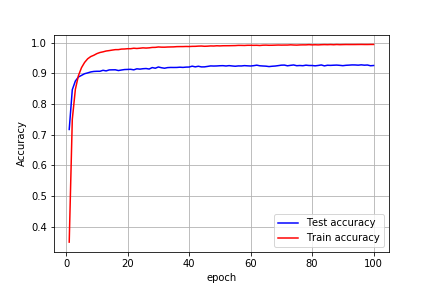
Training for 100 epochs without hyper-tuning the network I got the following results:
After tuning dropout, adding max-pooling layer and batch normalization I got the test accuracy of 99%.
So how I achieved more than 90% accuracy below different model with hyperparameter tuning.
As we had data in a format of train, test, dev. So we used test data to check the accuracy.
After testing we got these final results:
score = model.evaluate(finaltest,ytest , verbose=1)
print("Test loss:",score[0])
print("Test accuracy:",score[1])
7316/7316 [==============================] - 2s 251us/step
Test loss: 0.0511749743080253
Test accuracy: 0.9922886276653909
For testing, the same process needs to follow:
Pre-processing lowercase the sequence and taking the max sequence length of 100
Mapping char to index or numerical form
Using our model for the final prediction of a protein sequence of amino acid.
Since we have got a good result but it can get more improved
In order to get more accuracy model can be trained on a large data set.
By some more hyperparameter tuning, more accuracy can be achieved.
It can be used for other protein sequences classification problem by tuning or increasing the complexity of the model.
https://www.appliedaicourse.com/
https://www.biorxiv.org/content/10.1101/626507v3.full
https://arxiv.org/abs/1606.01781
https://www.kaggle.com/googleai/pfam-seed-random-split