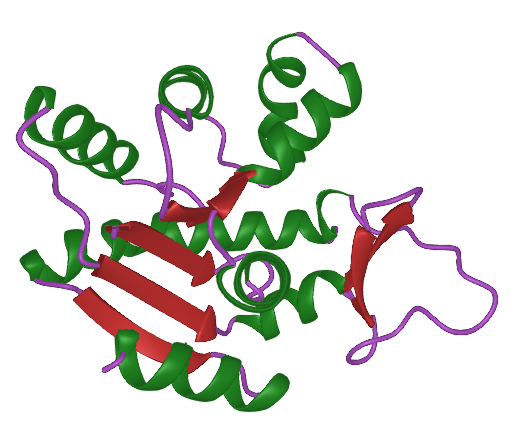
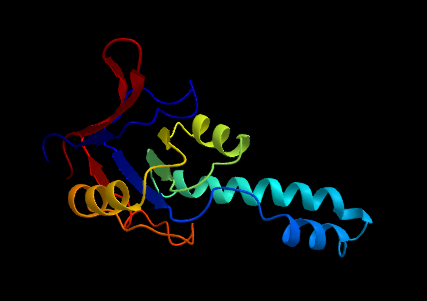
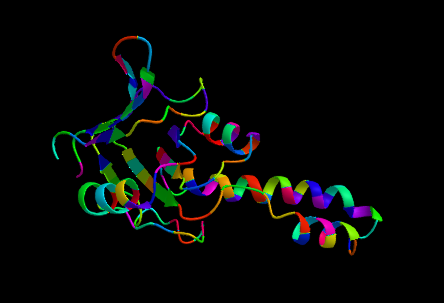
ProtPlot is a Julia package for rendering 3D protein ribbon using Makie.jl.
ProtPlot exports the Ribbon plot:
ribbon: makes a static render of the ribbon.ribbon!: renders the ribbon plot within an existing container (e.g.,SceneorAxis3).ribbon_scene: creates an interactive Scene to render the ribbon.
Ribbon plots are constructed from vectors of protein backbones represented as 3x3xL arrays.
For convenience, argument conversion methods are defined, and the ribbon constructors can take:
- a
3x3xLarray of residue backbone atom coordinates (N, Ca, C), or a vector of these for each chain. - a
BioStructures.MolecularStructureorBioStructures.Chain. - a PDB file path.
using GLMakie # use the GLMakie backend
using ProtPlot
# Create and display a ribbon plot in an interactive window
ribbon_scene("test/data/1ASS.pdb", backgroundcolor=:black, colormap=:jet)Use the colors keyword argument to customize colors at the residue level. This argument should be a vector of vectors, where each vector contains either:
- values between 0 and 1, representing the colors of each residue in their respective chains according to the
colormap. - a list of
Colorants, one per residue. For example,RGBA(1, 0, 0, 0.5)for a 50% transparent red. Load theColorTypesorColorspackage to create suchColorants.
# Load protein data from a PDB file
chains = readpdb("test/data/1ASS.pdb")
colors = rand.(length.(chains))
ribbon_scene(chains, colors=colors, colormap=:hsv)You may customize the geometry of the ribbon by specifying the value of attributes in the keyword arguments of your call. Here's a list of available attributes and their defaults:
-
secondary_structures = nothing(gets assigned by an algorithm by default; needs to be a vector ofVector{Int}where1means loop,2means helix, and3means strand) -
colors = nothing(gets assignedrange(0, 1, L)for each chain by default, mapping tocolormap; overrides colormap if colorants are given) -
colormap = :jet(see the ColorSchemes.jl catalogue; can also be a vector of colorants) -
coil_diameter = 0.4 -
coil_spline_quality = 20 -
coil_slice_quality = 20 -
helix_width = 2.0 -
helix_thickness = 0.5 -
helix_spline_quality = 20 -
helix_slice_quality = 20 -
strand_width = 2.0 -
strand_thickness = 0.5 -
strand_arrow_head_length = 5.0 -
strand_arrow_head_width = 3.5 -
strand_spline_quality = 20
Makie allows programmatic control over the camera.
Use the camcontrols keyword to control the initial view in a ribbon_scene call:
ribbon_scene("test/data/1ASS.pdb", camcontrols=(; lookat=Vec3f(30, 0, 60), eyeposition=Vec3f(160, -75, 0), upvector=Vec3f(0, 0, 1)))