This project aims to generate an enriched dataset from the PepBDB database for machine learning and computational biology research.
The script processes peptide-protein interaction data, extracts sequences, enriches them with various biochemical features, and creates a tabular dataset suitable for further analysis with random forests, XGBoost, etc. Each row is labeled as either a binding residue (1) or non-binding residue (0).
| AA | Protein Hydrophobicity | Protein Steric Parameter | Protein Volume | Protein Polarizability | Protein Helix Probability | Protein Beta Probability | Protein Isoelectric Point | Protein HSE Up | Protein HSE Down | Protein Pseudo Angles | Protein ASA | Protein Phi | Protein Psi | Protein SS H | Protein SS B | Protein SS E | Protein SS G | Protein SS I | Protein SS T | Protein SS S | Protein SS - | A | R | N | D | C | Q | E | G | H | I | L | K | M | F | P | S | T | W | Y | V | Binding Indices |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L | 0.6891891891891891 | 0.9607843137254901 | 0.8222778473091366 | 0.6241610738255033 | 0.6823529411764706 | 0.7473684210526315 | 0.40175219023779735 | 0.3333333333333333 | 0.42857142857142855 | 0.8699882132974325 | 0.4456686291000842 | 0.23066692000760028 | 0.0816007154035323 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.6666666666666666 | 0.0 | 0.0 | 0.0 | 0.16666666666666663 | 0.5555555555555556 | 1.0 | 0.0 | 0.5555555555555556 | 0.5 | 0.33333333333333326 | 0.0 | 0.0 | 0.25 | 0.30000000000000004 | 0.7142857142857142 | 0 |
| K | 0.518018018018018 | 0.6666666666666667 | 0.8610763454317898 | 0.7348993288590604 | 0.6941176470588235 | 0.25263157894736843 | 0.8723404255319149 | 0.0 | 0.5714285714285714 | 0.8747249797378288 | 0.7270984020185031 | 0.19703591107733237 | 0.09479096803040464 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.3333333333333333 | 0.4444444444444444 | 0.36363636363636365 | 0.30000000000000004 | 0.6666666666666666 | 0.5 | 0.3333333333333333 | 0.30000000000000004 | 0.5 | 0.2222222222222222 | 0.5 | 0.8571428571428571 | 0.4444444444444444 | 0.2 | 1.0 | 0.8 | 0.2857142857142857 | 0.125 | 0.2 | 0.42857142857142855 | 1 |
| D | 0.2072072072072072 | 0.7450980392156863 | 0.40175219023779735 | 0.3523489932885906 | 0.48235294117647065 | 0.18947368421052624 | 0.0 | 0.0 | 0.49999999999999994 | 0.6108288105124712 | 0.7711867992384177 | 0.1911457343720312 | 0.1553767046724793 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2222222222222222 | 0.4545454545454546 | 1.0 | 0.0 | 0.375 | 0.6666666666666666 | 0.2 | 0.5 | 0.0 | 0.0 | 0.42857142857142855 | 0.0 | 0.0 | 0.6666666666666666 | 0.6000000000000001 | 0.14285714285714285 | 0.0 | 0.09999999999999998 | 0.14285714285714285 | 1 |
peppi_data_imgs
├── binding
│ ├── img1.jpg
│ ├── img2.jpg
│ ├── img3.jpg
│ └── ...
└── nonbinding
├── img4.jpg
├── img5.jpg
├── img6.jpg
└── ...
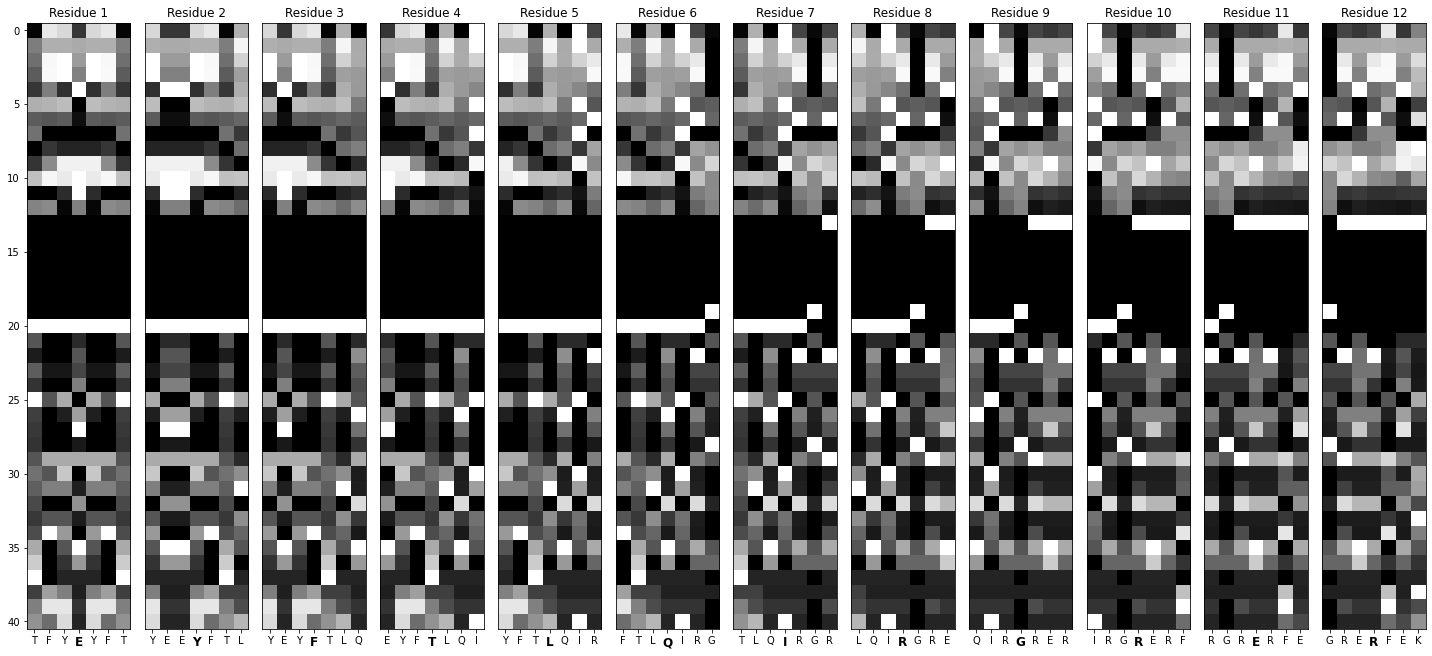 Note: the actual images have their dimensions flipped from what is shown. This image is magnified for illustrations purposes.
Note: the actual images have their dimensions flipped from what is shown. This image is magnified for illustrations purposes.
To run this script, you will need the following:
- Python 3.7+
- blast+
- mkdssp
- prodigy (
pip install prodigy-prot) - Python packages: pandas, numpy, biopython, scikit-learn
You can install the required Python packages using:
pip install pandas numpy biopython prodigy-prot scikit-learnYou can download the PepBDB dataset from here:
curl -O http://huanglab.phys.hust.edu.cn/pepbdb/db/download/pepbdb-20200318.tgzThe script begins by loading the peptidelist.txt file from the PepBDB database. The columns are renamed for better readability and convenience.
import pandas as pd
peptide_list = pd.read_csv(peptide_list_txt, sep='\s+', header=None)
headers = ['PDB ID', 'Peptide Chain ID', 'Peptide Length', 'Number of Atoms in Peptide',
'Protein Chain ID', 'Number of Atoms in Protein',
'Number of Atom Contacts', 'unknown1', 'unknown2', 'Resolution', 'Molecular Type']
peptide_list.columns = headersThe script filters out:
- Entries involving nucleic acids.
- Models with a resolution higher than 2.5 Å for quality.
- Peptides shorter than 10 amino acids.
peptide_list = peptide_list[peptide_list['Molecular Type'] != 'prot-nuc']
peptide_list = peptide_list[peptide_list['Resolution'] < 2.5]
peptide_list = peptide_list[peptide_list['Peptide Length'] >= 10]Sequences are extracted from PDB files using BioPython. We'll also filter out sequences containing non-standard amino acids.
from Bio.PDB import PDBParser
from Bio.SeqUtils import seq1
def extract_sequence(pdb_filename):
parser = PDBParser()
structure = parser.get_structure('X', pdb_filename)
for model in structure:
for chain in model:
residues = chain.get_residues()
sequence = ''.join(seq1(residue.get_resname()) for residue in residues if residue.get_resname() != 'HOH')
return sequence
peptide_list['Peptide Sequence'] = peptide_list['Peptide Path'].apply(extract_sequence)
peptide_list['Protein Sequence'] = peptide_list['Protein Path'].apply(extract_sequence)Using PRODIGY (with default parameters) to identify binding residues.
def label_residues(peptide_path, protein_path):
# Combine PDB files
with tempfile.NamedTemporaryFile(suffix='.pdb', delete=False) as temp_file:
output_path = temp_file.name
subprocess.run(['cat', peptide_path, protein_path], stdout=temp_file)
temp_file.close()
# Run PRODIGY
subprocess.run(['prodigy', '-q', '--contact_list', output_path])
ic_file_path = output_path.replace('.pdb', '.ic')
if not os.path.exists(ic_file_path):
raise FileNotFoundError(f"{ic_file_path} not found after running PRODIGY.")
contacts = pd.read_csv(ic_file_path, sep='\s+', header=None)
contacts.columns = ['peptide_residue', 'peptide_index', 'peptide_chain', 'protein_residue', 'protein_index', 'protein_chain']
peptide_binding_residues = sorted(contacts['peptide_index'].unique())
protein_binding_residues = sorted(contacts['protein_index'].unique())
os.remove(output_path)
return peptide_binding_residues, protein_binding_residues
peptide_list['Peptide Binding Indices'] = np.nan
peptide_list['Protein Binding Indices'] = np.nan
for index, row in peptide_list.iterrows():
peptide_binding_positions, protein_binding_positions = label_residues(row['Peptide Path'], row['Protein Path'])
peptide_list.at[index, 'Peptide Binding Indices'] = str(peptide_binding_positions)
peptide_list.at[index, 'Protein Binding Indices'] = str(protein_binding_positions)Using AAindex1 for residue-specific feature extraction.
from aaindex import *
features = {
'Hydrophobicity': hydrophobicity,
'Steric Parameter': steric_parameter,
'Volume': residue_volume,
'Polarizability': polarizability,
'Helix Probability': average_relative_probability_of_helix,
'Beta Probability': average_relative_probability_of_beta_sheet,
'Isoelectric Point': isoelectric_point
}
for feature_name, feature_function in features.items():
peptide_list[f'Peptide {feature_name}'] = peptide_list['Peptide Sequence'].apply(lambda x: feature_vector(x, feature_function))
peptide_list[f'Protein {feature_name}'] = peptide_list['Protein Sequence'].apply(lambda x: feature_vector(x, feature_function))Additional biochemical features are added, including HSE, ASA, DSSP codes, and PSSM profiles.
from helpers import safe_hse_and_dssp, one_hot_encode_row, extend_hse, get_pssm_profile
# Adding HSE, ASA, DSSP codes
peptide_list[['Peptide HSE Up', 'Peptide HSE Down', 'Peptide Pseudo Angles', 'Peptide SS', 'Peptide ASA', 'Peptide Phi', 'Peptide Psi']] = peptide_list['Peptide Path'].apply(lambda x: pd.Series(safe_hse_and_dssp(x)))
peptide_list[['Protein HSE Up', 'Protein HSE Down', 'Protein Pseudo Angles', 'Protein SS', 'Protein ASA', 'Protein Phi', 'Protein Psi']] = peptide_list['Protein Path'].apply(lambda x: pd.Series(safe_hse_and_dssp(x)))
# One-hot encoding DSSP codes
ss_columns = peptide_list.apply(one_hot_encode_row, axis=1)
peptide_list = pd.concat([peptide_list, ss_columns], axis=1)
# Extending HSE and Pseudo Angles to match peptide lengths
peptide_list['Protein HSE Up'] = peptide_list['Protein HSE Up'].apply(extend_hse)
peptide_list['Protein HSE Down'] = peptide_list['Protein HSE Down'].apply(extend_hse)
peptide_list['Protein Pseudo Angles'] = peptide_list['Protein Pseudo Angles'].apply(extend_hse)
peptide_list['Peptide HSE Up'] = peptide_list['Peptide HSE Up'].apply(extend_hse)
peptide_list['Peptide HSE Down'] = peptide_list['Peptide HSE Down'].apply(extend_hse)
peptide_list['Peptide Pseudo Angles'] = peptide_list['Peptide Pseudo Angles'].apply(extend_hse)
print('\033[1mHSE data extended...\033[0m')
# Adding PSSM profiles
peptide_list['Peptide PSSM'] = peptide_list['Peptide Sequence'].apply(get_pssm_profile)
peptide_list['Protein PSSM'] = peptide_list['Protein Sequence'].apply(get_pssm_profile)
print('\033[1mPSSMs generated...\033[0m')We'll also make life easier by reducing the dataset such that we only have one peptide or protein per row. Since PSI-BLAST can be tricky, it's also likely we'll have some empty alignments — we'll filter these out.
# Reducing to one peptide/protein per row
peptide_cols = [col for col in peptide_list.columns if 'Peptide' in col]
peptide_cols.remove('Peptide Length')
protein_cols = [col for col in peptide_list.columns if 'Protein' in col]
pep_data = peptide_list[peptide_cols]
pro_data = peptide_list[protein_cols]
combined_data = pd.DataFrame(np.vstack([pep_data, pro_data.values]))
combined_data.columns = pro_data.columns
peptide_list = combined_data
print(f'\033[1mBefore removing empty PSSMs, we have array shape of {peptide_list.shape}\033[0m')
contains_na = peptide_list['Protein PSSM'].apply(lambda df: df.empty or df.isna().any().any())
peptide_list = peptide_list[~contains_na]
peptide_list.reset_index(drop=True)
print(f'\033[1mRemoving empty PSSMs leads to array shape of {peptide_list.shape}\033[0m')
print('\033[1mData dimensions have been reduced...\033[0m')
print('\033[1mNow tabulating...\033[0m')
# Creating tabular dataset
list_of_feature_arrays = []
for i, row in peptide_list.iterrows():
arrs = make_tabular_dataset(row)
list_of_feature_arrays.append(arrs)
print(f'\r{i}/{peptide_list.shape[0]}', end='')
print('\033[1m\nConverted.\033[0m')
export = pd.concat(list_of_feature_arrays)
export = export.dropna()
export = export.reset_index(drop=True)
export.to_csv(peppi_data_csv, index=False)
print('\033[1mComplete!\033[0m')To run the script, simply execute:
tar -xzf pepbdb-20200318.tgz
python gendata.pygendata.py can also generate images in the style of the Visual dataset. To enable this option, set the --images flag to true and specify full paths for binding and non-binding images:
python gendata.py \
--images True \
--binding_path path/to/binding \
--nonbinding_path path/to/nonbindingIMPORTANT: Remember to modify paths.py with paths specific to your system.
Ensure you have the necessary input files and directories as specified in the script.
- The images directory
peppi_data_imgs.tgzand tabular datasetpeppi_data.csv.gzare NOT 1-1, and the CSV is not label file for the images. While they build from the same data, they do not contain the same number of records.- 811,830 records in
peppi_data.csv- Binding: 110,268
- Non-binding: 701,562
- 806,129 images in
peppi_data_imgs- Binding: 109,880
- Non-binding: 696,249
- 811,830 records in
- This is because some rows (residues) in
peppi_data.csvhave NaN values. Before exporting the CSV, these rows alone are simply dropped. However, that same faulty row/reside can appear in several images (since each image is a representation of seven contiguous resides). To maintain usability, ALL images containing that resiude are dropped.
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J. (1997) “Gapped BLAST and PSI-BLAST: a new generation of protein database search programs.” Nucleic Acids Res. 25:3389-3402. PubMed
- Kabsch, W., & Sander, C. (1983). Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577-2637. PMID: 6667333
- Kawashima, S., Ogata, H., & Kanehisa, M. (1999). AAindex: amino acid index database. Nucleic Acids Res., 27, 368-369. PMID: 9847231
- Kawashima, S., & Kanehisa, M. (2000). AAindex: amino acid index database. Nucleic Acids Res., 28, 374. PMID: 10592278
- Nakai, K., Kidera, A., & Kanehisa, M. (1988). Cluster analysis of amino acid indices for prediction of protein structure and function. Protein Eng., 2, 93-100. PMID: 3244698
- Tomii, K., & Kanehisa, M. (1996). Analysis of amino acid indices and mutation matrices for sequence comparison and structure prediction of proteins. Protein Eng., 9, 27-36. PMID: 9053899
- Touw, W.G., et al. (2015). A series of PDB related databases for everyday needs. Nucleic Acids Research, 43(Database issue), D364-D368.
- Wen, Z., He, J., Tao, H., & Huang, S.-Y. (2018). PepBDB: a comprehensive structural database of biological peptide–protein interactions. Bioinformatics, 35(1), 175–177. https://doi.org/10.1093/bioinformatics/bty579