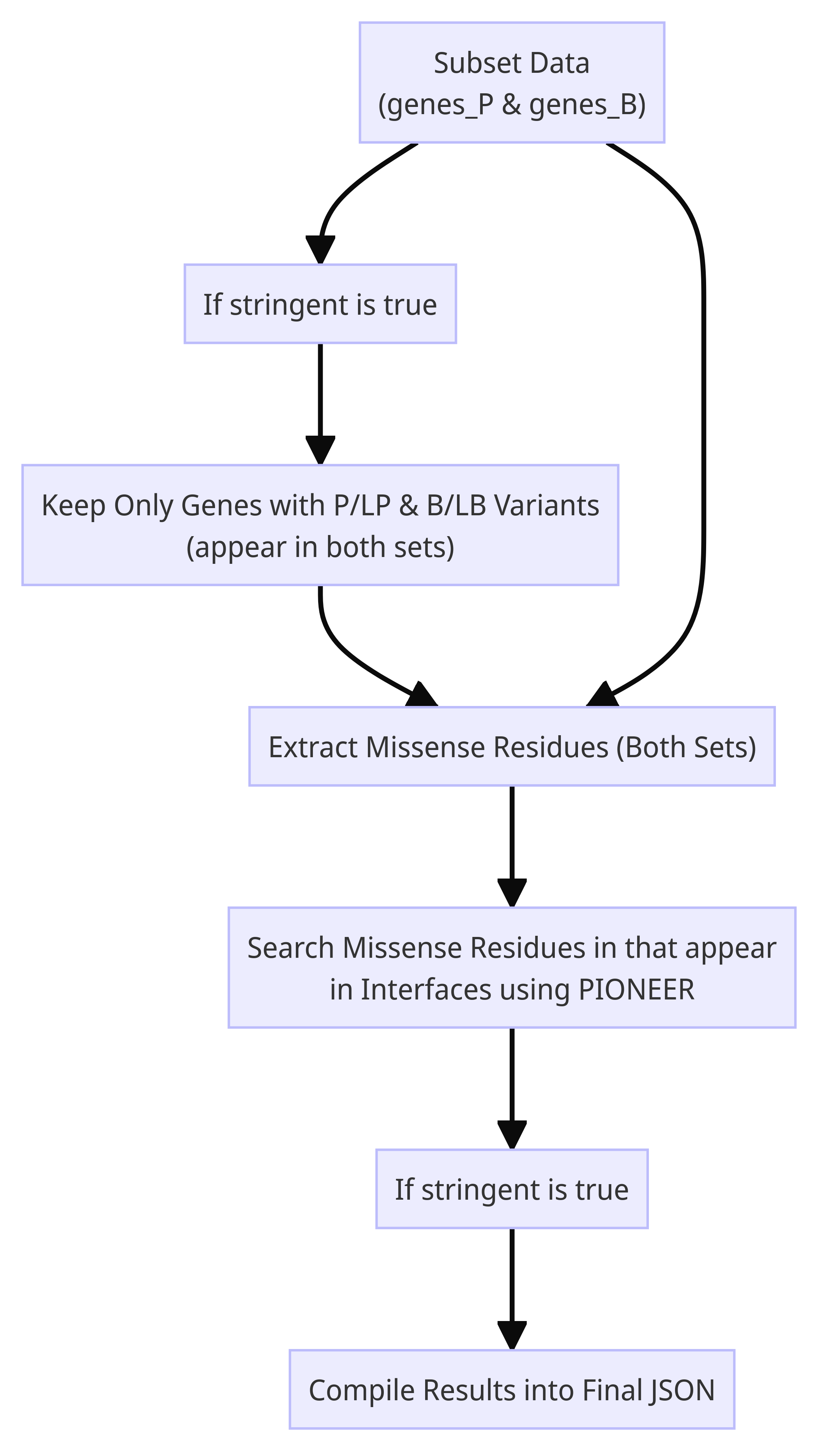
The shell script helps you find known variant missense residues (P/LP and B/LB) that occur at protein-protein interfaces for use as in silico controls. It currently accepts search output data from ClinVar as inputs and returns JSON information filtered out from PIONEER.
All subscripts can be accessed independently and used from the command line as separate utilities:
python {script}.py <arg1> <arg2> ...- Python 3.x
- Unipressed (
pip install unipressed) - Human interactome data from PIONEER, as TSV. (You can use
data.shto download this for you.)
To execute the script, open your terminal and navigate to the folder containing the script. Then run:
./vici.sh \
-B [path/to/benign/variant/table] \
-P [path/to/pathogenic/variant/table] \
-O [output_folder_name]For example, if you have benign variant data in a file named benign_data.tsv, pathogenic variant data in pathogenic_data.tsv, and you want to output to a folder named results, run:
./vici.sh -B benign_data.tsv -P pathogenic_data.tsv -O resultsThe script accepts the following command-line options:
-
-B, --benign: Specifies the path to the benign variant table from Clinvar. This is a required argument. -
-P, --pathogenic: Specifies the path to the pathogenic variant table from Clinvar. This is a required argument. -
-O, --output: Specifies the name of the output folder where the results will be saved. This is a required argument. -
-h, --help: Displays a help message detailing how to use the script and what each option does. -
-s, --stringent: If flag is set to true, the pipeline will return a final JSON contaning only those proteins that have both P/LP AND B/LB variants that occur at interfaces.
To run the script with specific benign and pathogenic variant tables and an output folder named analysis_output:
./vici.sh -B benign_variants.tsv -P pathogenic_variants.tsv -O analysis_outputTo display the help menu:
./vici.sh -h- Missense residues JSONs (for both TSVs).
- Missense residues appearing at interfaces for both JSONs.
- A combined JSON including data from both P/LP and B/LB interface residue JSONs.
- Directories containing TSVs subset from input tables, based on gene name.
All output files will be saved in the specified output_folder.
subset.py: This script is used to subset a TSV file by the values in its second column, which is assumed to be a gene name. The output is a directory of TSV files, each corresponding to a gene name.common.py: (Optional) This script is used to remove files from two folders that don't have a shared basename. Shared basenames tell us that genes in question have both benign and pathogenic variants.rois.py: The residues of interest script is used to extract numbers from protein change codes in TSV files. The output is a JSON file containing a dictionary of lists, where each key is the basename of a TSV file and each value is a missense residue position. We'll search against these numbers to see if they appear in an interface.search.py: Find residues in the interface of a protein-protein interaction that are also missense variants in a gene. The output is a JSON file containing a dictionary of dictionaries, where each key is a gene name and each value is a dictionary of dictionaries, where each key is a partner gene name and each value is a list of common residues.shared.py: (Optional) Finds the shared keys between two JSON files and write them to a new JSON file. For control purposes, it tells you which proteins have both P/LP & B/LP variant residues in their interfaces, and which proteins those interfaces are with.
- Implement more detailed error reporting and logging.
- Add support for variant data from gnomAD.