List of participants and affiliations:
- Chaoran Chen, ETH Zurich, Basel Switzerland (Team Leader)
- Adrian Lison, ETH Zurich, Basel Switzerland
- Auguste Rimaite, ETH Zurich, Basel Switzerland
- Jennifer Chang, Fred Hutchinson Cancer Research Center
- Theo Sanderson, Francis Crick Institute
- Alex Kramer, UC Santa Cruz
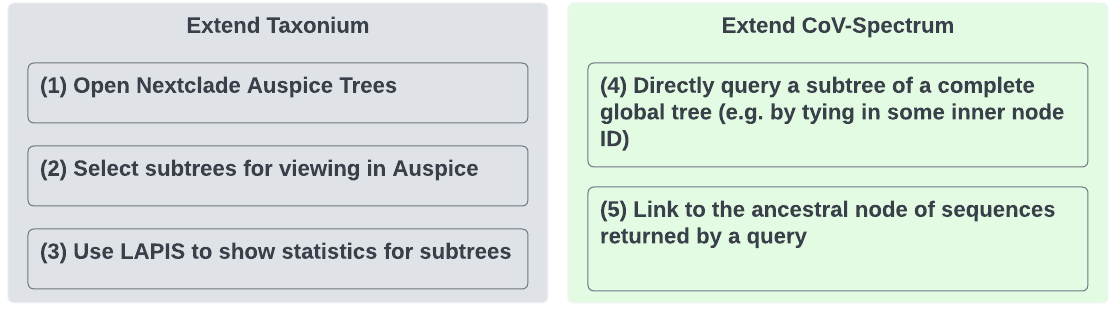
Extend Taxonium
Extend CoV-Spectrum
Link to the CoV-Spectrum instance during the codeathon
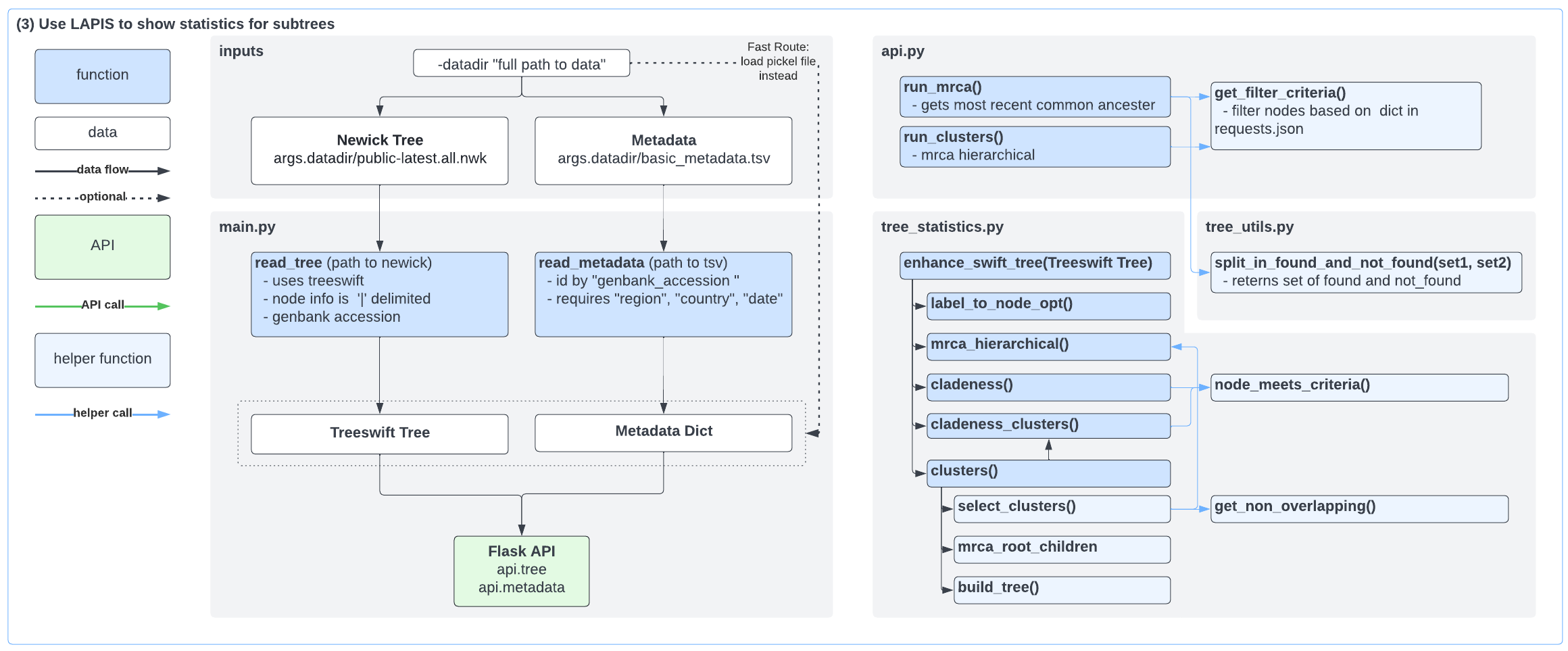
New Cladeness API
curl --header "Content-Type: application/json" \
--request POST \
--data '["ON875035", "ON861689"]' \
https://cladeness.cov-spectrum.org/mrca
We created an API endpoint to compute a statistic we call cladeness, which is defined as the percentage of a clade occupied by a set of samples, where the clade is that at the MRCA of the set of samples.
We added a Tree Statistics panel to CoV-Spectrum that computes the MRCA of the selected sample set, and displays the cladeness and size of clusters within the MRCA subtree.
In each cluster above, users can click the button to open the ancestor node in Taxonium (displaying the global SARS-CoV-2 tree)
Taxonium can now load tree files in the Nextstrain JSON (v2) format.
Example Nextstrain JSON file (monkeypox)
Trees can be loaded by file upload or by providing a URL.
Example tree displayed in Taxonium, with metadata and mutations parsed from Nextstrain file:
Taxonium can now export subtrees to Nextstrain JSON format.
In server-side mode (for example at cov2tree.org) Taxonium can directly load a subtree into Nextstrain by clicking View clade in Nextstrain. In local mode (when uploading a tree file from your computer or a URL), the Nextstrain JSON for a subtree can be downloaded (Download Nextstrain JSON) and then loaded into auspice.us for visualization.
In Taxonium's global SARS-CoV-2 instance (cov2tree.org), clades in CoVSpectrum can now be viewed by clicking Find this clade in CovSpectrum at a given node.
- Chen, C., Nadeau, S., Yared, M., Voinov, P., Xie, N., Roemer, C. and Stadler, T., 2022. CoV-Spectrum: analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics, 38(6), pp.1735-1737.
- Sanderson, T., 2022. Taxonium: a web-based tool for exploring large phylogenetic trees. bioRxiv.
- Chen, C, Roemer, C. and Stadler, T., 2022. LAPIS is a fast web API for massive open virus sequencing databases. arXiv.
- Huddleston, J., Hadfield, J., Sibley, T.R., Lee, J., Fay, K., Ilcisin, M., Harkins, E., Bedford, T., Neher, R.A. and Hodcroft, E.B., 2021. Augur: a bioinformatics toolkit for phylogenetic analyses of human pathogens. Journal of open source software, 6(57).
- Hadfield, J., Megill, C., Bell, S.M., Huddleston, J., Potter, B., Callender, C., Sagulenko, P., Bedford, T. and Neher, R.A., 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics, 34(23), pp.4121-4123.
- Moshiri, N., 2020. TreeSwift: A massively scalable Python tree package. SoftwareX, 11, p.100436.
- Aksamentov, I., Roemer, C., Hodcroft, E.B. and Neher, R.A., 2021. Nextclade: clade assignment, mutation calling and quality control for viral genomes. Journal of Open Source Software, 6(67), p.3773.
- Kramer, A., Turakhia, Y. and Corbett-Detig, R., 2021. ShUShER: private browser-based placement of sensitive genome samples on phylogenetic trees. Journal of Open Source Software, 6(66), p.3677.