SRA Metadata is rich, sometimes too rich. There's no single standardized location for many types of useful information. This is a generalizable framework for pulling SRA metadata and summarizing usefully, with some common use-cases ready to go.
Even the type of study is heterogeneous for different submitters. The location of for instance tissue and developmental stage specifications within SRA metadata also varies, as do the values used to describe particular tissues.
Third-party efforts to load all metadata into a relational db suggest how widespread the problem is-- a customizable localized implementation helps many.
Requires python3 and python packages noted in requirements.txt
To get started, git clone to subdirectory MetadataTable, and create a working subdirectory named "testing"
cd testing
source bin/activate
pip3 install -r ../MetadataTable/requirements.txt
python3 ../MetadataTable/metadatatable.py -h
Please note that this a minimal implementation, quickly written; very few informative warning messages if problems are encountered
Retrieve all RNA-Seq datasets for taxon 7955, save output as a tab-delimited version :
python3 ../MetadataTable/metadatatable.py -e <your_email> -t "(biomol_transcript[properties] OR study_type_transcriptome_analysis[properties] OR strategy_rna_seq[properties] OR strategy_FL_cDNA[properties]) AND txid7955[Organism]" -ot 7955.tsv
>python metadatatable.py -h
usage: MetadataTable [-h] [-ox OUTPUT_XML] [-ot OUTPUT_TSV] [-i INPUT_XML]
[-e EMAIL] [-t TERM [TERM ...]] [-u] [-f] [-x XPATH]
optional arguments:
-h, --help show this help message and exit
-ox OUTPUT_XML, --output_xml OUTPUT_XML
Path for saving xml file
-ot OUTPUT_TSV, --output_tsv OUTPUT_TSV
Path for saving parsed file
-i INPUT_XML, --input_xml INPUT_XML
Path to input file
-e EMAIL, --email EMAIL
Let NCBI know who you are (required)
-t TERM [TERM ...], --term TERM [TERM ...]
Query terms
-u, --unlimited Retrieve unlimited records
-f, --full Whether to output full table (Only for builtin template)
-x XPATH, --xpath XPATH
Path to a csv file for xpath query
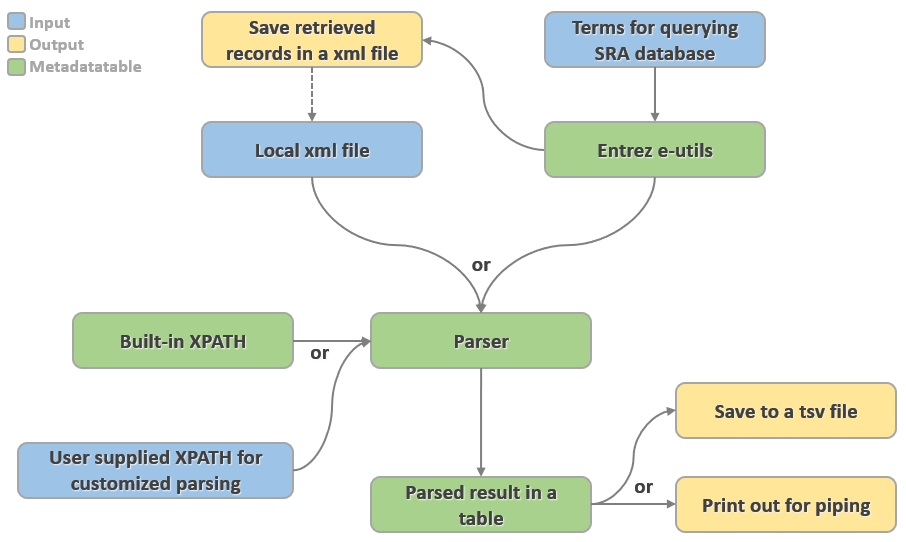
Input mode
Users can choose which input mode they want. These two modes cannot be used together.
-i input.xml
Use a xml file as input.
-e <your_email> -t searchterm
Query Entrez using the terms specified by -t.
Output mode
Users can save both xml and tsv file, if filenames are given. xml output can be large, it can be convenient to have a local copy while exploring which summary fields are informative.
(blank)
If no output mode specified, metadatatable will print out the parsed
results.
-ox output.xml
Save downloaded records in a xml file.
-ot output.tsv
Save parsed records in a tsv file.
Retrieval
-u
Retrieve unlimited records. If not used, metadatatable will abort if
the results from querying Entrez are more then 100,000
Parsing
-f
For built-in parsing template only. Output the full table. It will
be silently ignored if using user supplied xpath file.
-x xpath.csv
If supplied, metadatatable will use read the xpath query from the
specified csv file, and -c and -f will be silently ignored.
-c case
Use built-in parsing template. Choose from (rnaseq, source). Default
is rnaseq. Note that "source" template is not implemented yet. This
argument currently is ignored.
Tara Tufano
Lukas Wagner
Yadi Zhou