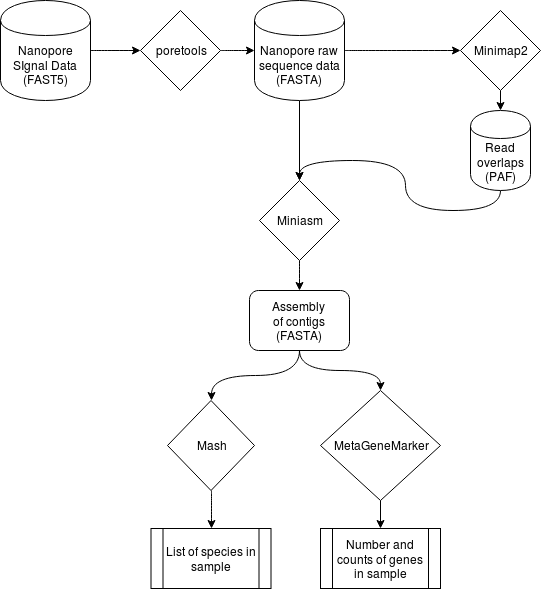
The world's simplest open-source nanopore alignment and assembly pipeline. This is meant to be an educational tool that Just Works(TM), and walks a user through assembling a metagenome from raw nanopore reads. This pipeline also performs gene annotation using MetaGeneMark and gives the user taxanomic information through Mash.
- Educational tool for teaching undergrads or high school students the basics of genome assembly
- Simple tool for WIMP (What's In My Pot) analysis that doesn't require any external tools
- Citizen scientist that wants to sequence whatever's on their roommate's toothbrush
Reads are first classified based on genomic distance using 4mers using Mash. The reads are classified into bins which are then assembled into genomes using canu. The resulting assemblies are then blasted using TAXBLAST to see the taxanomic information from the sample. The pipeline also does simple gene annotation using MetaGeneMark.
Pipeline must be run on a computer with atleast 4 GB of RAM
dockergit
Install the Docker Community Edition here for your distribution.
Run docker application Preferences -> Advanced Set Memory slider to the highest settings Apply changes
$ git clone https://github.com/NCBI-Hackathons/SpoonFedNanopore.git
$ cd SpoonFedNanopore/
$ docker build .
$ docker images
# Use the hash given by this command, and replace YOURHASH with the Image ID given
$ docker run -it -p 8888:8888 -v $PWD:/work YOURHASH
TODO instruction for building from docker store
Current workflow only implements the Canu assembly of raw reads and then the taxonomic information. Later steps will introduce the read clustering by Mash.
- Li, H. (2016). Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32(14), 2103–2110.
- Li, H. (2017). Minimap2: pairwise alignment for nucleotide sequences. arXiv.
- Loman, N. J. and Quinlan, A. R. (2014). Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics 30(23), 3399–3401.
- Loman, N. J., Quick, J. and Simpson, J. T. (2015). A complete bacterial genome assembled de novo using only nanopore sequencing data. Nature Methods 12(8), 733–735.