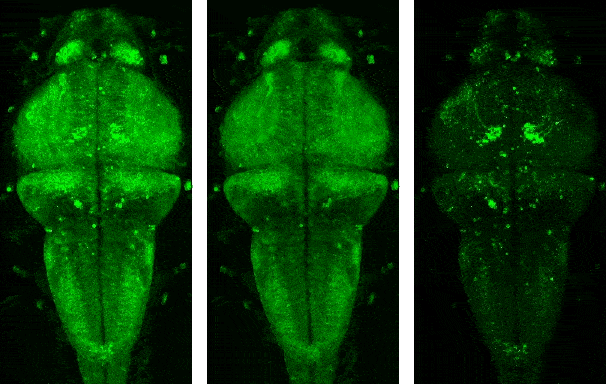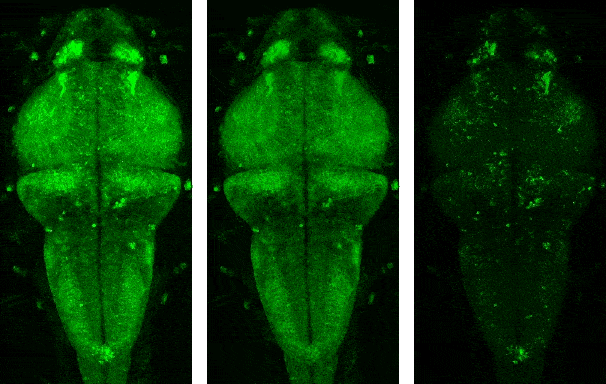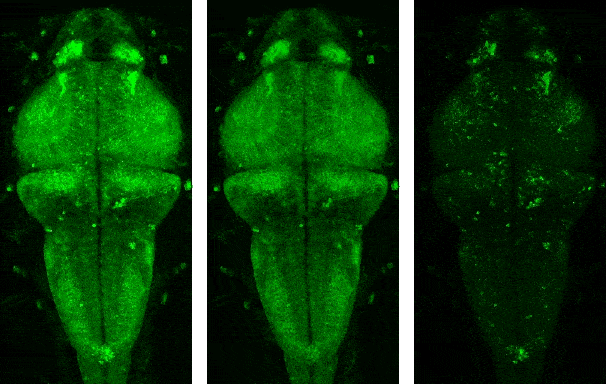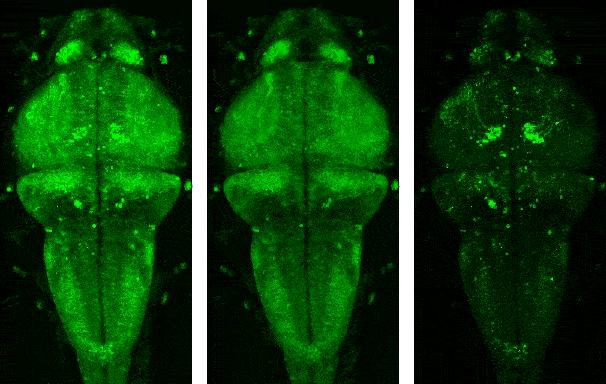
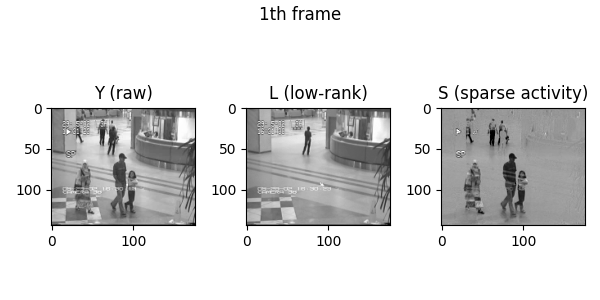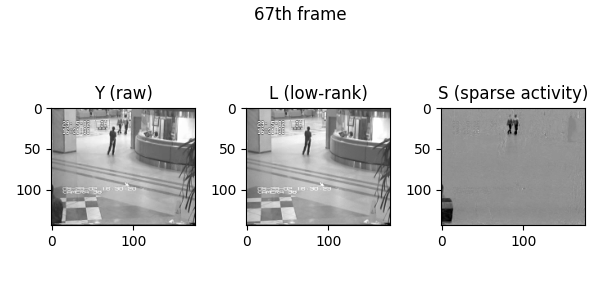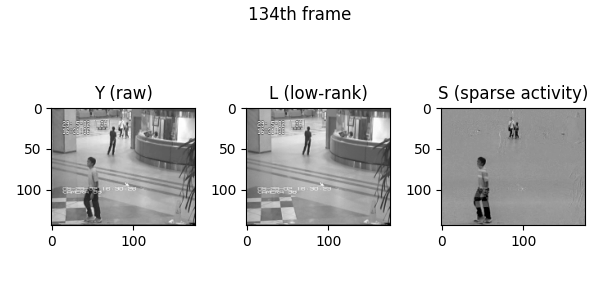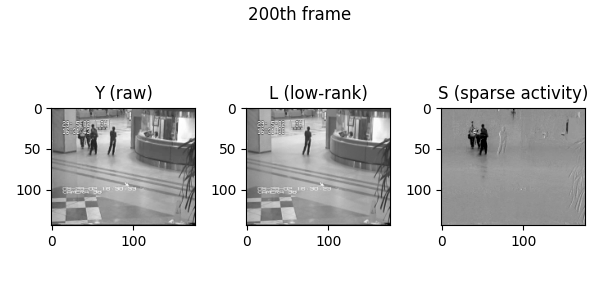
Demo videos acquired using BEAR. Input video, low-rank component, and sparse component from left to right.
Official source codes for "Efficient neural network approximation of robust PCA for automated analysis of calcium imaging data", MICCAI 2021 [1].
There is also Matlab implementation (~16MB) of BEAR.
Source codes for PCP, IALM, and GreGoDec are from lrslibrary. Source codes for OMWRPCA are from onlineRPCA. (Both last visited on March, 2021.)
In demo.ipynb notebook, you can follow how to use BEAR for surveillance video. There is a video saved in this notebook, where they may not visible if you simply open the notebook in github. One way to view all things in the notebook is opening it with VS Code.
Following is the visualization result in the notebook. You can see that low rank L and sparse S is well decomposed from input video Y.
You can run BEAR by very simple code. Just download or clone this repository and use the scripts below.
--D refers the name of the data and --d enables the default settings of several hyperparameters.
There are already two videos in data folder: small surveillance video, and calcium imaging data of mouse which are widely used in RPCA paper. You can first try with these data. You can try BEAR with as follows:
python scripts/run_BEAR.py --D hall --d True
python scripts/run_BEAR.py --D demoMovie --d TrueAlso you can download additional calcium imaging data of zebrafish in this Google Drive (~25MB). If you want to see more examples, try these. Download, unzip, and move .tif and .mat files inside the data folder. Then, you can try BEAR with as follows:
python scripts/run_BEAR.py --D confocal_zebrafish --d True
python scripts/run_BEAR.py --D confocal_zebrafish_2 --d TrueResults will be automatically saved in results/BEAR/(name of the data)_(Y/L/S).
Hardware specification
OS : Ubuntu 18.0.4
CPU : Intel i7-9700K
GPU : GeForce RTX 2080 Ti 11GB
RAM : 128GMRequirements (May be okay with different versions.)
torch==1.7.0
numpy==1.19.4
psutil==5.7.2
scipy==1.5.3
scikit-image==0.17.2
mat73==0.46Calcium imaging data of zebrafish we have used in paper is in this Google Drive (~10GB). They are very large to show that BEAR is fast and scalable to use in large calcium imaging data.
- Figure 3. Phase diagram.
python scripts/phase_diagram.py --D None --d None- Table 1. and Figure 4. Decomposition of the zebrafish caclium imaging data.
python scripts/run_Greedy_BEAR.py --D zebrafish_150 --d True
python scripts/run_Greedy_BEAR.py --D zebrafish_1000 --d TrueDue to the size of data, loading files itself does take long time (Minutes in HDD). And for 1000 length video, about 100GB of RAM is required.
- Figure 5. and Figure 6. Cascaded BEAR for analysis of neuronal activity.
python scripts/run_Cascaded_BEAR.py --D demoMovie --d True
python scripts/run_Cascaded_BEAR.py --D spinning_confocal --d TrueFor accuracy, number of epochs in default settings of Cascaded BEAR is very large.
Can be observed that loss value does not decrease actually after small number of epochs.
You can reduce the args.epoch if you want.
If you find this repository or our paper helpful, please cite our paper:
@inproceedings{han2021efficient,
title={Efficient neural network approximation of robust pca for automated analysis of calcium imaging data},
author={Han, Seungjae and Cho, Eun-Seo and Park, Inkyu and Shin, Kijung and Yoon, Young-Gyu},
booktitle={International Conference on Medical Image Computing and Computer-Assisted Intervention},
pages={595--604},
year={2021},
organization={Springer}
}