DCASE2023: FEW-SHOT BIOACOUSTIC EVENT DETECTION USING BEATS, ADAPTIVE FRAME-SHIFTS AND SPECTRAL GATING
💥 A PIPELINE FOR FINE-TUNING BEATs ON ESC50 DATASET IS PROVIDED HERE. The rest of the repository is on training a prototypical network using BEATs as feature extractor
⬇️
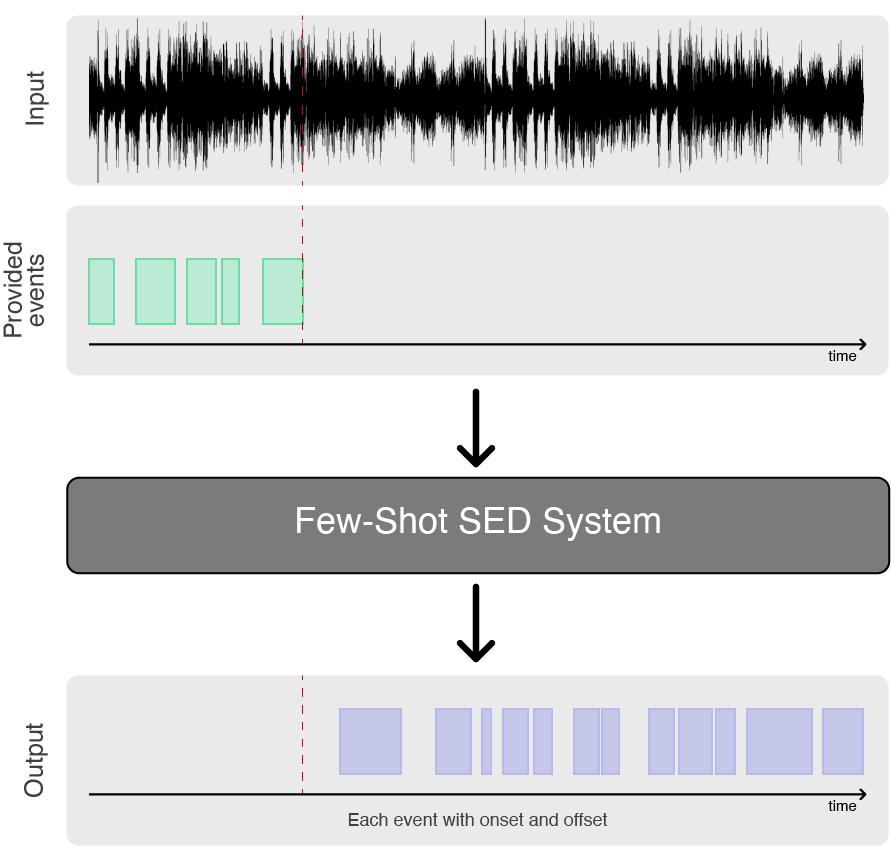
Few-shot learning is a highly promising paradigm for sound event detection. It is also an extremely good fit to the needs of users in bioacoustics, in which increasingly large acoustic datasets commonly need to be labelled for events of an identified category (e.g. species or call-type), even though this category might not be known in other datasets or have any yet-known label. While satisfying user needs, this will also benchmark few-shot learning for the wider domain of sound event detection (SED).
Few-shot learning describes tasks in which an algorithm must make predictions given only a few instances of each class, contrary to standard supervised learning paradigm. The main objective is to find reliable algorithms that are capable of dealing with data sparsity, class imbalance and noisy/busy environments. Few-shot learning is usually studied using N-way-K-shot classification, where N denotes the number of classes and K the number of examples for each class.
Text in this section is borrowed from c4dm/dcase-few-shot-bioacoustic
Our contribution:
This repository is the result of our submission to the DCASE2023 challenge task5: Few-shot Bioacoustic Event Detection. It containts the necessary code to train a prototypical network with BEATs as feature extractor on the data given by the DCASE challenge.
This repository's main objective is to keep active to tackle future DCASE challenges, if you wish to help us improve this repository / collaborate with us, please do not hesitate to send us a message!
Requirements
In this section are listed the requirements. Note that we make extensive use of Docker for easier reproducibility.
Setup
We have made a small wrapper to download the DCASE data and the BEATs model. Only the base folder needs to be specified:
./dcase_setup.sh /BASE/FOLDER/The script should create a DCASE folder containing all the DCASE data (i.e. Development and Evaluation set) and a BEATs folder containing the model weights in the specified base folder.
Once the necessary files have been dowloaded, you can either pull the Docker image and rename it:
docker pull docker pull ghcr.io/ninanor/rare_species_detections:main
docker tag docker pull ghcr.io/ninanor/rare_species_detections:main beatsOr create the Docker image from the Dockerfile located in our repository:
git clone https://github.com/NINAnor/rare_species_detections.git
cd rare_species_detections
docker build -t beats -f Dockerfile .Processing the data
First we need to process the DCASE data (i.e. denoising, resampling ... and saving the data as numpy array). For this we can use:
./preprocess_data.sh /BASE/FOLDERThe script will create a new folder DCASEfewshot containing three subfolders (train, validate and evaluate). Each of these folder contains the processed data in the form of numpy arrays.
Train the model
It is now possible to train the network using prototypicalbeats/trainer.py:
./train_model.sh /BASE/FOLDERThe training script should create a log folder in the base folder (lightning_logs/) in which the model weights (version_X/checkpoints/*.ckpt) and the training configuration (version_X/checkpoints/config.yaml) are stored.
Using the model on the Validation / Evaluation dataset
To run the prediction use the script test_model. Note that the file CONFIG.yaml file need to be updated. In particular you will need to change the model_path, status (either test or validate). and set_type (Validation_Set or Evaluation_Set)
./test_model.sh /BASE/FOLDERtest_model.sh creates a result file eval_out.csv in the BASE/FOLDER containing all the detections made the model.
Note that there are other advanced options. For instance, if --wav_save is specified, the script will also return a .wav file for all files containing additional channels: the ground truth labels, the predicted labels, the distance to the POS prototype and finally the p-values. The .wav file can be opened in Audacity to be inspected more closely.
Computing the resulting metrics
Once the eval_out.csv has been created, it is possible to get the results for our approach. Note that the metrics can only be computed for the Validation_Set as it contains all ground truth labels as opposed to the Evaluation_Set for which only the 5 first samples of the POS class are labelled.
docker run -v $CODE_DIR:/app \
-v $DATA_DIR:/data \
--gpus all \
beats \
poetry run python evaluation/evaluation_metrics/evaluation.py \
-pred_file /data/eval_out.csv \
-ref_files_path /data/DCASE/Development_Set_annotations/Validation_Set \
-team_name BEATs \
-dataset VAL \
-savepath /data/.The results we obtained:
Evaluation for: BEATs VAL
BUK1_20181011_001004.wav {'TP': 15, 'FP': 35, 'FN': 16, 'total_n_pos_events': 31}
BUK1_20181013_023504.wav {'TP': 2, 'FP': 258, 'FN': 22, 'total_n_pos_events': 24}
BUK4_20161011_000804.wav {'TP': 1, 'FP': 30, 'FN': 46, 'total_n_pos_events': 47}
BUK4_20171022_004304a.wav {'TP': 7, 'FP': 17, 'FN': 10, 'total_n_pos_events': 17}
BUK5_20161101_002104a.wav {'TP': 31, 'FP': 7, 'FN': 57, 'total_n_pos_events': 88}
BUK5_20180921_015906a.wav {'TP': 4, 'FP': 24, 'FN': 19, 'total_n_pos_events': 23}
ME1.wav {'TP': 9, 'FP': 18, 'FN': 2, 'total_n_pos_events': 11}
ME2.wav {'TP': 41, 'FP': 27, 'FN': 0, 'total_n_pos_events': 41}
R4_cleaned recording_13-10-17.wav {'TP': 19, 'FP': 14, 'FN': 0, 'total_n_pos_events': 19}
R4_cleaned recording_16-10-17.wav {'TP': 30, 'FP': 8, 'FN': 0, 'total_n_pos_events': 30}
R4_cleaned recording_17-10-17.wav {'TP': 36, 'FP': 9, 'FN': 0, 'total_n_pos_events': 36}
R4_cleaned recording_TEL_19-10-17.wav {'TP': 52, 'FP': 12, 'FN': 2, 'total_n_pos_events': 54}
R4_cleaned recording_TEL_20-10-17.wav {'TP': 64, 'FP': 8, 'FN': 0, 'total_n_pos_events': 64}
R4_cleaned recording_TEL_23-10-17.wav {'TP': 84, 'FP': 8, 'FN': 0, 'total_n_pos_events': 84}
R4_cleaned recording_TEL_24-10-17.wav {'TP': 99, 'FP': 14, 'FN': 0, 'total_n_pos_events': 99}
R4_cleaned recording_TEL_25-10-17.wav {'TP': 99, 'FP': 9, 'FN': 0, 'total_n_pos_events': 99}
file_423_487.wav {'TP': 57, 'FP': 13, 'FN': 0, 'total_n_pos_events': 57}
file_97_113.wav {'TP': 11, 'FP': 27, 'FN': 109, 'total_n_pos_events': 120}
Overall_scores: {'precision': 0.2911279078300433, 'recall': 0.4938446186969832, 'fmeasure (percentage)': 36.631}
Taking the idea further:
- Computing the mahalanobis distance instead of the Euclidean distance
- Implementing a p-value filtering to detect outlier distances from the prototypes
Acknowlegment and contact
For bug reports please use the issues section.
For other inquiries please contact Benjamin Cretois or Femke Gelderblom
Cite this work
Gelderblom, F., Cretois, B., Johnsen, P., Remonato, F., & Reinen, T. A. (2023). Few-shot bioacoustic event detection using beats. Technical report, DCASE2023 Challenge.