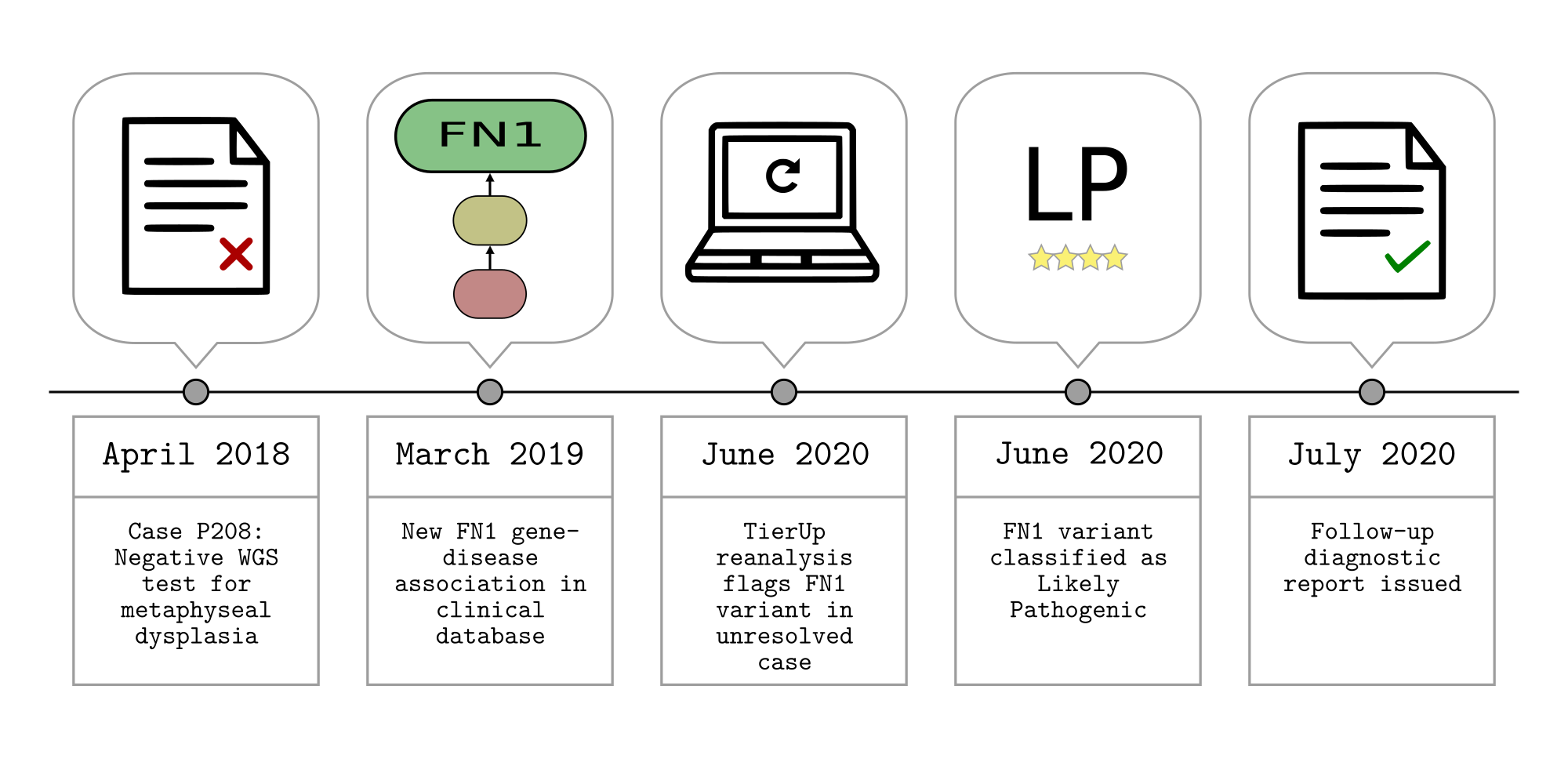
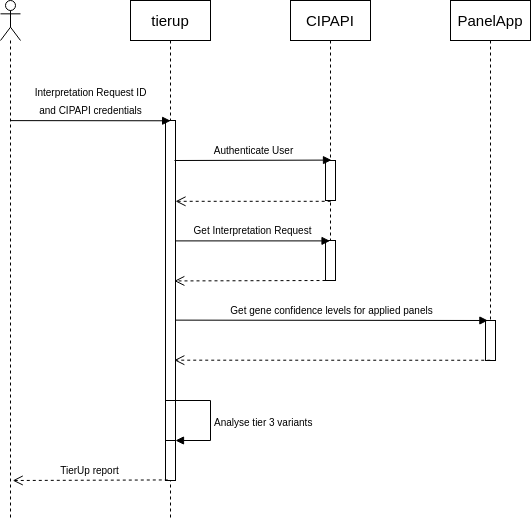
Reanalysis of unresolved rare disease cases using TierUp version 0.3.0.
- db.csv - Merged TierUp results for 948 anonymised cases
- Metadata parsed from TierUp input files using custom scripts:
- cohort.csv - Number of patients recruited per case
- rd_group_referrals.csv - Rare disease group referrals from all cases
TierUp takes GEL interpretation request JSON files as input. These files contain variants prioritised by the GEL tiering pipeline during the initial analysis. TierUp can also download the input files given a case identifier, provided the user is connected to the NHS Health and Social Care Network and has valid GeL CIP-API credentials.
We processed all unresolved cases using the following command:
time parallel tierup -c config.ini -j {} ::: *.jsons &> results.txt
This created *.tierup.csv files with the results for each case. Additionally, results.txt reported the time taken to process 948 cases on this machine:
real 76m43.785s
user 140m17.023s
sys 5m1.144s
We combied TierUp reanalysis results files into a single CSV file using csvstack. Patient identifiers were pseudonymed and any fields that were not required for publication were dropped.
Descriptive statistics are calculated from db.csv and accompanying files as recorded in the analysis notebook.
The /files directory contains figures and data that were produced manually.