In our Institutional Review Board (IRB) approved retrospective study, we gathered a large collection of MRI scans of the pancreas, which included both T1 and T2 weighted images, sourced from multiple centers with varying MRI protocols imposing a heterogeneity challenge. We create a new deep learning tool, named PANSegNet, to identify pancreatic tissue in these scans. We also used publicly available CT scans to test our algorthm for pancreas segmentation. We obtained the state of the art results both in CT and MRI scans.
Our data is the largest ever MRI pancreas dataset so far in in the literature and can be found here (PanSegData): https://osf.io/kysnj/. Please cite our work when you use our code and data.
Abstract:
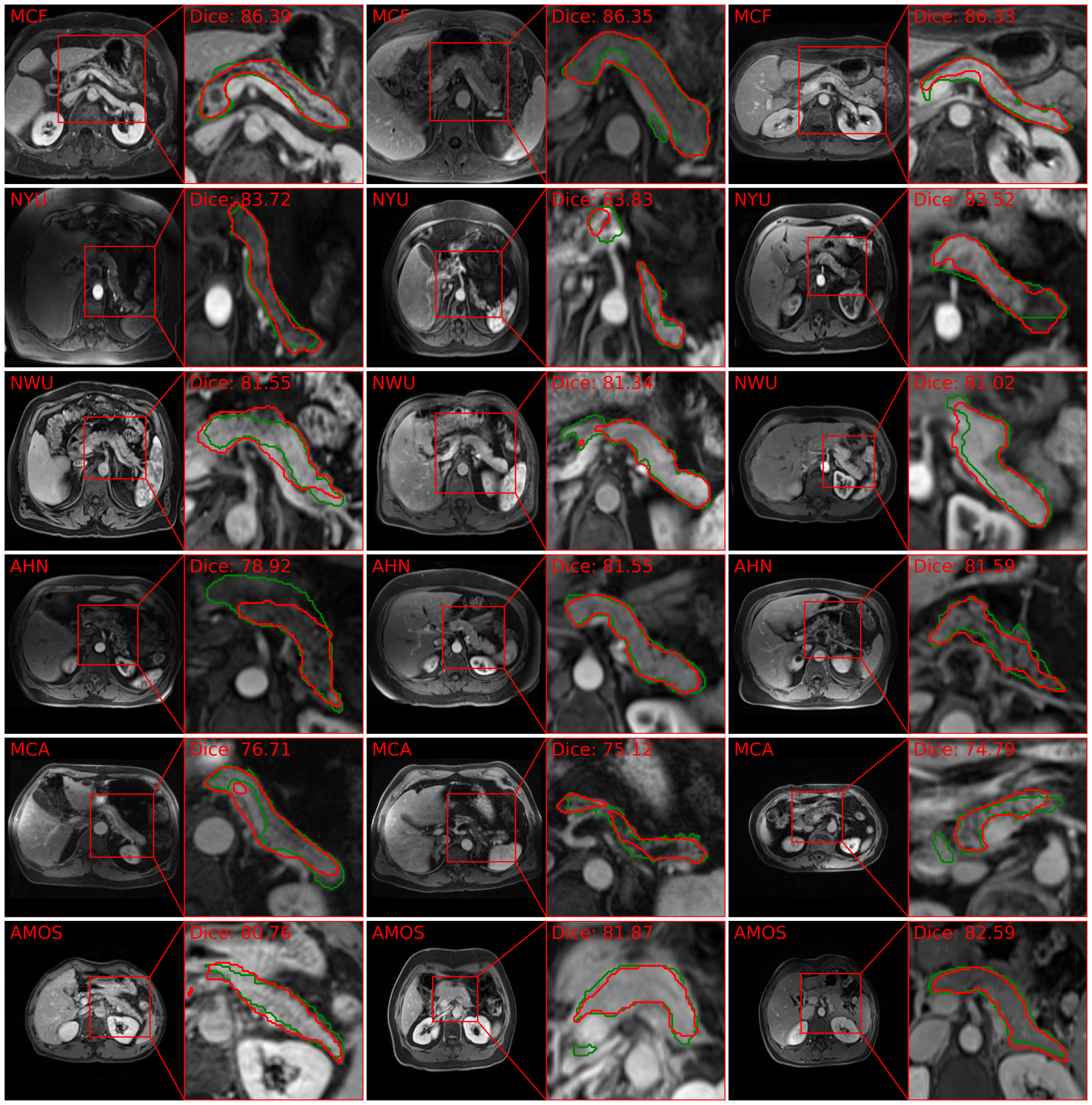
Automated volumetric segmentation of the pancreas on cross-sectional imaging is needed for diagnosis and follow-up of pancreatic diseases. While CT-based pancreatic segmentation is more established, MRI-based segmentation methods are understudied, largely due to a lack of publicly available datasets, benchmarking research efforts, and domain-specific deep learning methods. In this retrospective study, we collected a large dataset (767 scans from 499 participants) of T1-weighted (T1W) and T2-weighted (T2W) abdominal MRI series from five centers between March 2004 and November 2022. We also collected CT scans of 1,350 patients from publicly available sources for benchmarking purposes. We developed a new pancreas segmentation method, called \textit{PanSegNet}, combining the strengths of \textit{nnUNet} and a \textit{Transformer} network with a new linear attention module enabling volumetric computation. We tested \textit{PanSegNet}’s accuracy in cross-modality (a total of 2,117 scans) and cross-center settings with Dice and Hausdorff distance (HD95) evaluation metrics. We used Cohen’s kappa statistics for intra and inter-rater agreement evaluation and paired t-tests for volume and Dice comparisons, respectively. For segmentation accuracy, we achieved Dice coefficients of 88.3% (± 7.2%, at case level) with CT, 85.0% (± 7.9%) with T1W MRI, and 86.3% (± 6.4%) with T2W MRI. There was a high correlation for pancreas volume prediction with
Here we provide the segmentation dataset public.
https://osf.io/kysnj/. Our proposed model achieved accurate segmentation on large scale pancreas MRI datasets.
Our model is based on nnUNet v1 and please follow the environment setup correspondingly. We setup our training and model following standard nnUNet style in "nnTransUNetTrainerV2". Therefore, all training and inference can be easily follow the standard nnUNet training and inference.
For example, in order to run inference on several target centers like A, B, C, D for T1 scans, you can use followings script directly. Please note, each center should be organized in standard nnUNet inference style, which means the original scans should be in nii.gz format within one folder and file name should end with _0000 for detection.
# Add your target domain in the domain_list
# Add the base directory where the target domain is stored
domain_list=(A B C D )
base_dir="XXX"
for domain in ${domain_list[@]}
do
echo "Inference on ${domain}"
input_dir="${base_dir}/${domain}/imagesTs"
mkdir ${base_dir}/${domain}/inference_segmentation
OUTPUT_DIRECTORY="${base_dir}/${domain}/inference_segmentation"
CUDA_VISIBLE_DEVICES=0 nnUNet_predict -tr nnTransUNetTrainerV2 \
-i ${input_dir} -o ${OUTPUT_DIRECTORY} -t 110 -m 3d_fullres --folds 0
# CUDA_VISIBLE_DEVICES=0 nnUNet_evaluate_folder -ref ${base_dir}/${domain}/labelsTr -pred ${OUTPUT_DIRECTORY} -l 1
doneWe also provide the public model Pretrained Weight for easy access. Note that CT model is marked with ID 109, MRI T1 model is marked with 110, and MRI T2 image is marked with 111. Please be careful for environment variable setups and follow the guidance of nnUNet. Also, other comparision model's pretrained weights are provided in huggingface
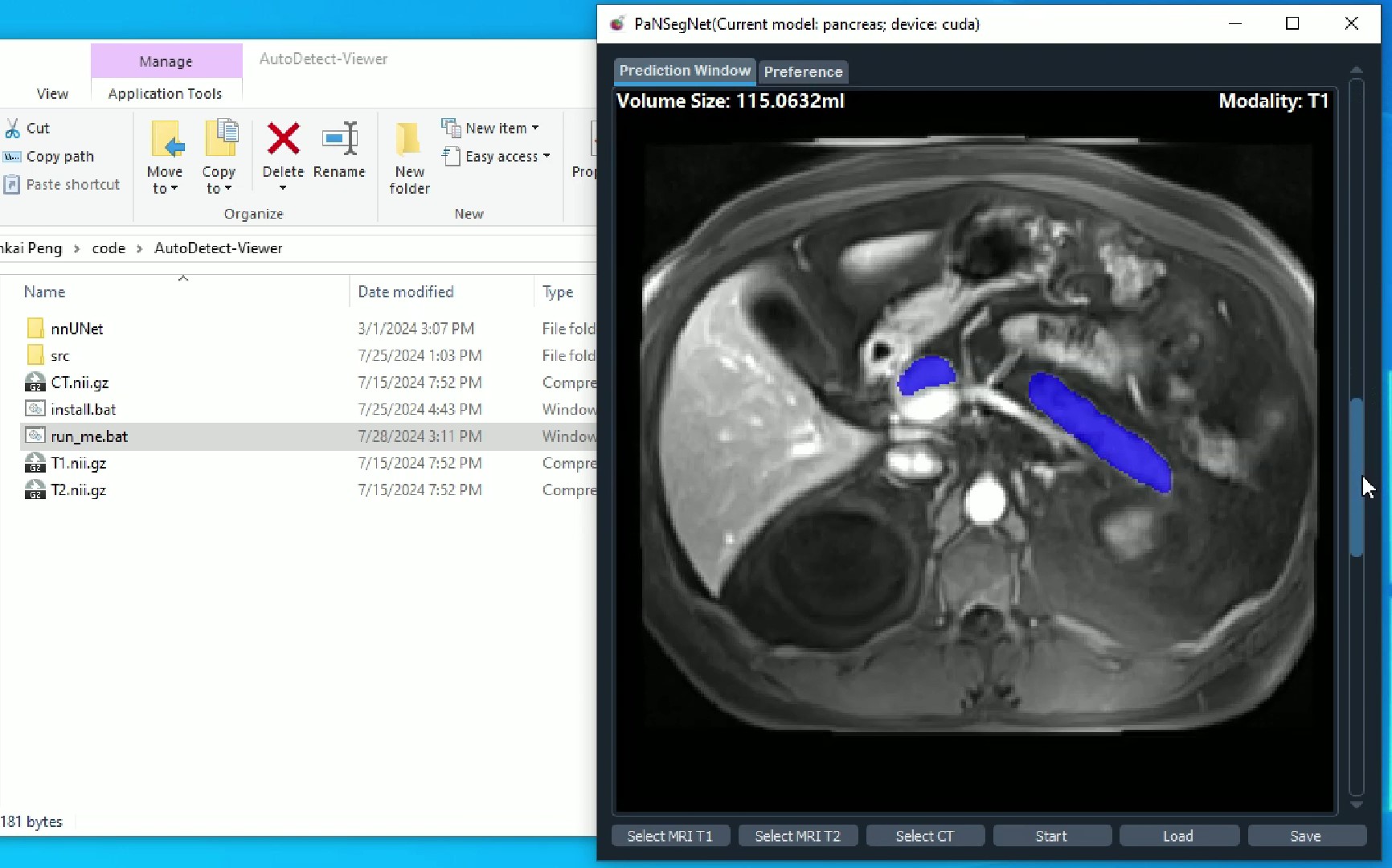
Additionally, we provide one intuitive GUI software for running the segmentation with "all-in-one" format. Please check the details PaNSegNet
Please cite our work if you find it is helpful
@article{zhang2024large,
title={Large-Scale Multi-Center CT and MRI Segmentation of Pancreas with Deep Learning},
author={Zhang, Zheyuan and Keles, Elif and Durak, Gorkem and Taktak, Yavuz and Susladkar, Onkar and Gorade, Vandan and Jha, Debesh and Ormeci, Asli C and Medetalibeyoglu, Alpay and Yao, Lanhong and others},
journal={arXiv preprint arXiv:2405.12367},
year={2024}
}