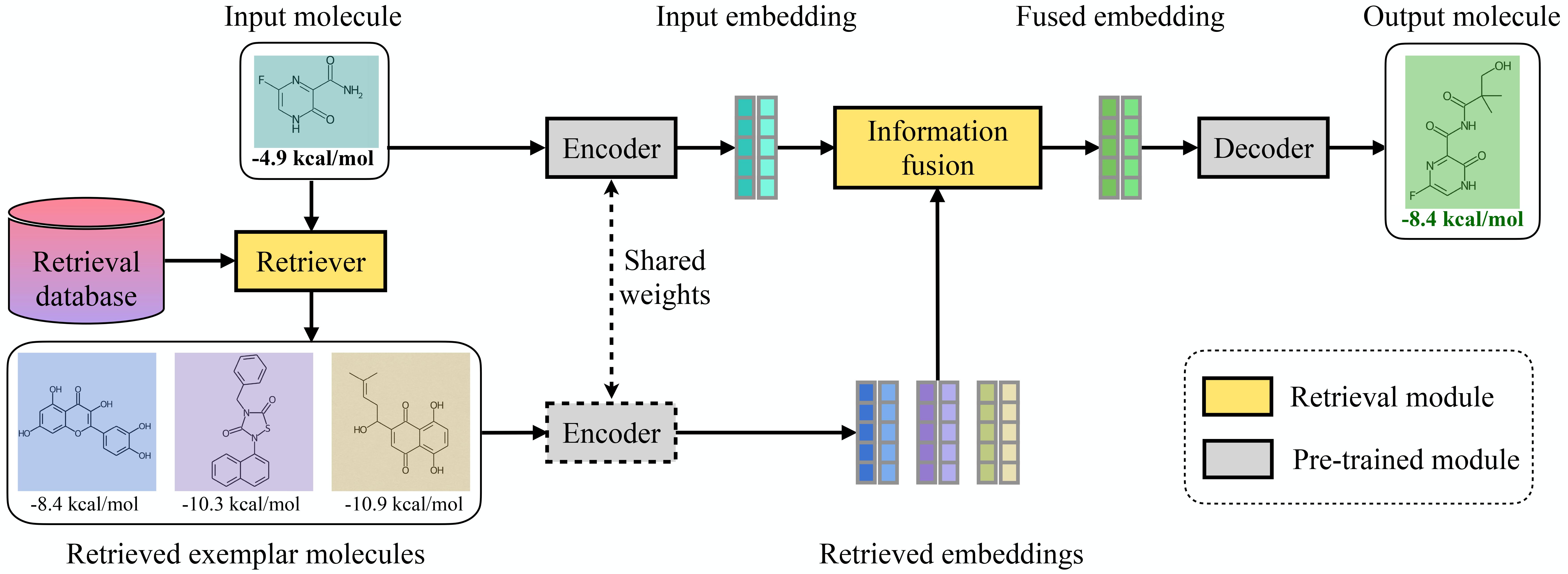
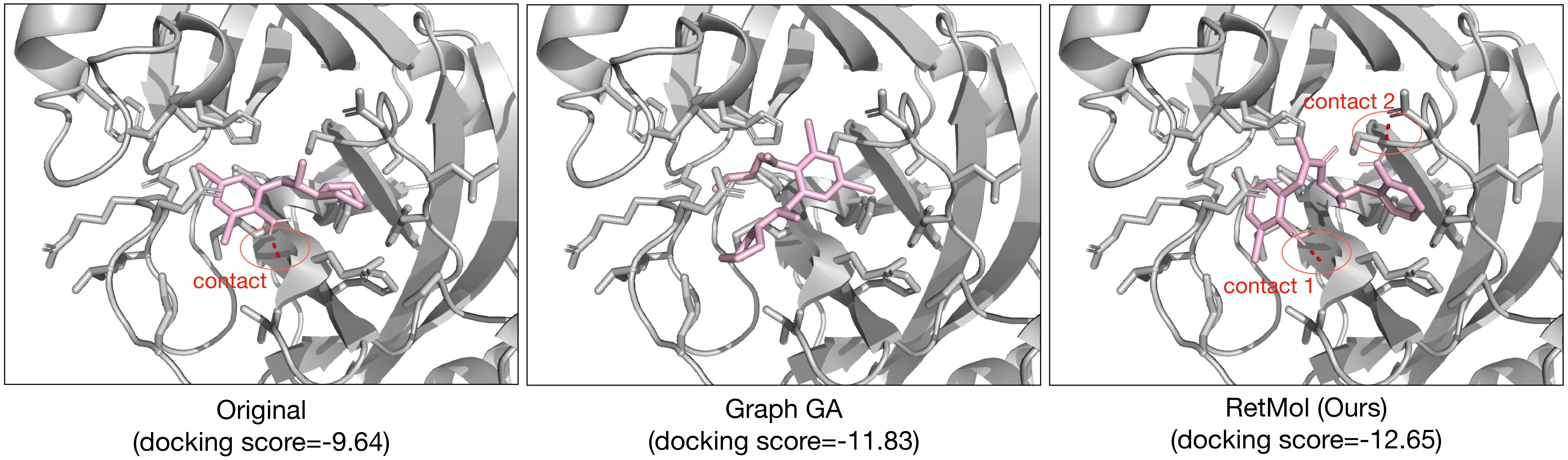
This repo contains codes used in the ICLR 2023 paper:
Retrieval-based Controllable Molecule Generation.
Abstract: Generating new molecules with specified chemical and biological properties via generative models has emerged as a promising direction for drug discovery. However, existing methods require extensive training/fine-tuning with a large dataset, often unavailable in real-world generation tasks. In this work, we propose a new retrieval-based framework for controllable molecule generation. We use a small set of exemplar molecules, i.e., those that (partially) satisfy the design criteria, to steer the pre-trained generative model towards synthesizing molecules that satisfy the given design criteria. We design a retrieval mechanism that retrieves and fuses the exemplar molecules with the input molecule, which is trained by a new self-supervised objective that predicts the nearest neighbor of the input molecule. We also propose an iterative refinement process to dynamically update the generated molecules and retrieval database for better generalization. Our approach is agnostic to the choice of generative models and requires no task-specific fine-tuning. On various tasks ranging from simple design criteria to a challenging real-world scenario for designing lead compounds that bind to the SARS-CoV-2 main protease, we demonstrate our approach extrapolates well beyond the retrieval database, and achieves better performance and wider applicability than previous methods.
The easiest way to run the code is via the provided docker image. The docker image has several conda environments that enable experimentations without much further configurations.
- clone this repo. assume its directory path on your machine is
$RetMol_dir - pull the docker image via
docker pull moonlightlane/retmol - start a docker container with the command
docker run --gpus all -it --name retmol -p 8888:8888 -v $RetMol_dir:/mnt/retmol --shm-size 8G moonlightlane/retmol:latest /bin/bash. Replace$RetMol_dirwith this repo's directory path. after this step, you will be inside the docker environment. - run
conda activate drug. Then runexport PROJECT_HOME=/mnt/retmol, and finallycd /mnt/retmol. - the container can be exited via
exitand re-entered viadocker start $IDthendocker exec -it $ID /bin/bash, where$IDis the identifier number of the docker container. This identifier can be found viadocker container ls -all
After these steps, you are almost ready to try out a bunch of things (see below). You may need to download certain files; please refer to each section below for the necessary additional steps.
MolBARTcontains the training scripts and the model filesinferencecontains scripts that run various experiments including QED, penalized logP, gsk3+jnk3, and SARS-CoV-2guacamolcontains scripts that run the guacamol experimenteval_notebookscontains jupyter notebooks to reproduce results in our paperdownload_scriptscontains scripts to download necessary files for training, inference, and reproducing results
The model checkpoints can be downloaded via cd download_scripts then sh download_models.sh . The RetMol checkpoints are now in /mnt/retmol/models/retmol_* (assume this step is done inside the docker container)
- Download the result files. First
cd download_scripts, thensh download_results_reproduce.sh. This will create a directoryresults_reproduceunder$PROJECT_HOME. - Go to the folder
eval_notebooks. There are three jupyter notebooks that reproduces plots and visualizations that appear in our paper. These can be run by first running this command inside Dockerjupyter notebook --ip 0.0.0.0 --no-browser --allow-rootand then open the link in your browser.
- Training leverages the pre-computed molecule similarity data, which can be downloaded by first
cd download_scriptsand thensh download_data_ret_precompute.sh. This will create a directorydata/similarity-precomputeunder$PROJECT_HOME. - run
bash train_megatron_retrieval.shto train on the ZINC dataset, orbash train_megatron_retrieval_chembl.shto train on the chebl dataset.
Note that this training only involves a very lightweight information fusion module (< %5 of the total number of model parameters; see Section 2.1 and Appendix A for further details). The rest of the model parameters (e.g., encoder, decoder) are not trained and remain frozen.
- Download the necessary pre-computed similarity and embedding files by
cd download_scriptsand thensh download_data_ret_precompute.sh. This step can be skipped if it has already been done in the section "run training" above - run
export MASTER_PORT=1234 cd inference- each of the experiments can be run as following:
- for QED experiment:
python run_qed.py --model_path models/retmol_zinc --similarity_thres 0.6 --chunk 1 - For penalized logP experiment:
python run_logp-sa.py --model_path models/retmol_zinc --similarity_thres 0.6 --chunk 1 - for gsk3-jnk3 experiment:
python run_gsk3jnk3.py --model_path models/retmol_chembl --test_data_path data/gsk3jnk3/inference_input/inference_inputs.csv --ret_data_path chembl-4attr --ret_mode per-itr-faissNote that for the first two experiments (QED and penalized logP), the commands above runs for one-eighth (100) of all input molecules (800). The last option,--chunk, with options ranging from 1 to 8, changes which portions of the input molecules to run.
- for QED experiment:
- Download the necessary files by
cd download_scriptsand thendownload_data_guacamol.sh - run the experiment by
python run_retrieval_ga.py --benchmark_id 1. Change the--benchmark_idto run different guacamol benchmarks,
- Get Autodock-GPU:
- under
$PROJECT_HOME, clone the Autodock-GPU repo:git clone https://github.com/ccsb-scripps/AutoDock-GPU.git cd AutoDock-GPUexport GPU_INCLUDE_PATH=$YOUR_CUDA_PATH/include; thenexport GPU_LIBRARY_PATH=$YOUR_CUDA_PATH/lib64. You need to identify$YOUR_CUDA_PATH; for example,$YOUR_CUDA_PATH=/usr/local/cuda-10.2.make DEVICE=GPU NUMWI=256. You can change the last number to some smaller number, i.e.,128or64.- Note: the above steps are necessary because autodock-gpu is built specific to the system's CUDA version; pre-built binary that does not match your system's CUDA version will not work
- under
- Download the necessary files via
cd download_scriptsandsh download_data_cov.sh - Run the experiment via
cd inferenceandpython run_cov.py --model_path models/retmol_chembl --sim_thres 0.6
This repo in part utilizes codes from the following:
- https://catalog.ngc.nvidia.com/orgs/nvidia/teams/clara/models/megamolbart
- https://github.com/NVIDIA/cheminformatics
- https://github.com/MolecularAI/MolBART
- https://github.com/wengong-jin/multiobj-rationale
- https://github.com/wengong-jin/icml18-jtnn
- https://github.com/BenevolentAI/guacamol
This work is made available under the Nvidia Source Code License-NC. Please check the LICENSE file.
The model checkpoints are shared under CC-BY-NC-SA-4.0. If you remix, transform, or build upon the material, you must distribute your contributions under the same license as the original.
This work may be used non-commercially, meaning for research or evaluation purposes only. For business inquiries, please contact researchinquiries@nvidia.com.
Please cite out work if you find it useful:
@inproceedings{retmol2023iclr,
author = {{Wang}, Zichao and {Nie}, Weili and {Qiao}, Zhuoran and {Xiao}, Chaowei and {Baraniuk}, Richard and {Anandkumar}, Anima},
title = {Retrieval-based Controllable Molecule Generation},
booktitle = {International conference on learning representations (ICLR)},
year = {2023}
}