The coronavirus package provides a tidy format dataset of the 2019 Novel Coronavirus COVID-19 (2019-nCoV) epidemic. The raw data pulled from the Johns Hopkins University Center for Systems Science and Engineering (JHU CCSE) Coronavirus repository.
More details available
here, and a csv format
of the package dataset available
here
Source: Centers for Disease Control and Prevention’s Public Health Image Library
As this an ongoing situation, frequent changes in the data format may occur, please visit the package news to get updates about those changes
Install the CRAN version:
install.packages("coronavirus")Install the Github version (refreshed on a daily bases):
# install.packages("devtools")
devtools::install_github("RamiKrispin/coronavirus")While the coronavirus CRAN
version is updated
every month or two, the Github (Dev)
version is updated on a
daily bases. The update_dataset function enables to overcome this gap
and keep the installed version with the most recent data available on
the Github version:
library(coronavirus)
update_dataset()Note: must restart the R session to have the updates available
Alternatively, you can pull the data using the
Covid19R project data
standard
format
with the refresh_coronavirus_jhu function:
covid19_df <- refresh_coronavirus_jhu()
head(covid19_df)
#> date location location_type location_code location_code_type data_type value lat long
#> 1 2020-06-03 Afghanistan country AF iso_3166_2 cases_new 758 33.93911 67.709953
#> 2 2020-06-07 Afghanistan country AF iso_3166_2 recovered_new 45 33.93911 67.709953
#> 3 2020-06-02 Afghanistan country AF iso_3166_2 cases_new 759 33.93911 67.709953
#> 4 2020-06-04 Afghanistan country AF iso_3166_2 cases_new 787 33.93911 67.709953
#> 5 2020-06-08 Afghanistan country AF iso_3166_2 recovered_new 296 33.93911 67.709953
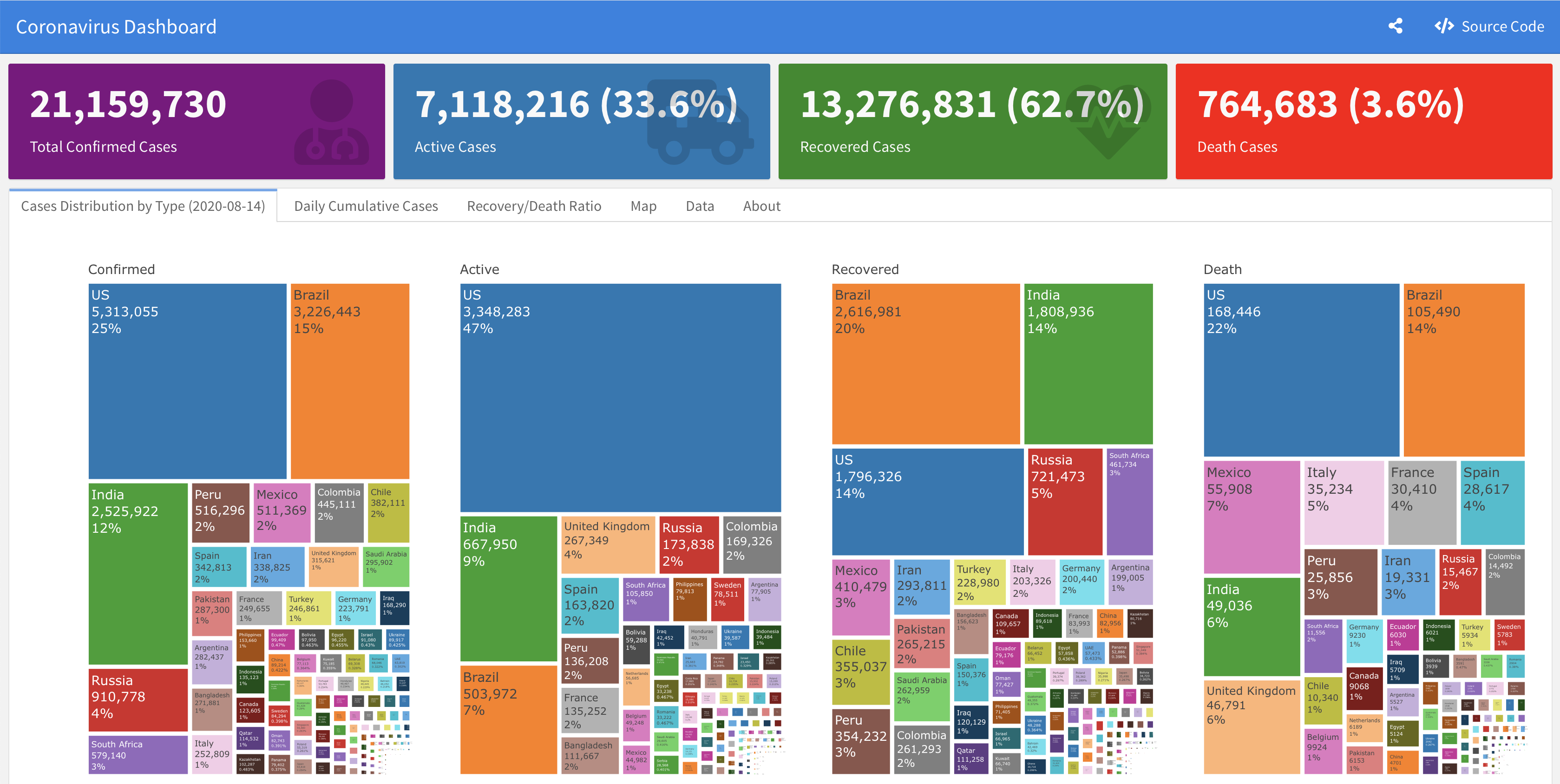
#> 6 2020-06-06 Afghanistan country AF iso_3166_2 recovered_new 68 33.93911 67.709953A supporting dashboard is available here
data("coronavirus")This coronavirus dataset has the following fields:
date- The date of the summaryprovince- The province or state, when applicablecountry- The country or region namelat- Latitude pointlong- Longitude pointtype- the type of case (i.e., confirmed, death)cases- the number of daily cases (corresponding to the case type)
head(coronavirus)
#> date province country lat long type cases
#> 1 2020-01-22 Afghanistan 33.93911 67.709953 confirmed 0
#> 2 2020-01-23 Afghanistan 33.93911 67.709953 confirmed 0
#> 3 2020-01-24 Afghanistan 33.93911 67.709953 confirmed 0
#> 4 2020-01-25 Afghanistan 33.93911 67.709953 confirmed 0
#> 5 2020-01-26 Afghanistan 33.93911 67.709953 confirmed 0
#> 6 2020-01-27 Afghanistan 33.93911 67.709953 confirmed 0Summary of the total confrimed cases by country (top 20):
library(dplyr)
summary_df <- coronavirus %>%
filter(type == "confirmed") %>%
group_by(country) %>%
summarise(total_cases = sum(cases)) %>%
arrange(-total_cases)
summary_df %>% head(20)
#> # A tibble: 20 x 2
#> country total_cases
#> <chr> <int>
#> 1 US 5141208
#> 2 Brazil 3057470
#> 3 India 2329638
#> 4 Russia 895691
#> 5 South Africa 566109
#> 6 Mexico 492522
#> 7 Peru 483133
#> 8 Colombia 410453
#> 9 Chile 376616
#> 10 Iran 331189
#> 11 Spain 326612
#> 12 United Kingdom 313394
#> 13 Saudi Arabia 291468
#> 14 Pakistan 285191
#> 15 Bangladesh 263503
#> 16 Argentina 260911
#> 17 Italy 251237
#> 18 Turkey 243180
#> 19 France 239355
#> 20 Germany 219540Summary of new cases during the past 24 hours by country and type (as of 2020-08-11):
library(tidyr)
coronavirus %>%
filter(date == max(date)) %>%
select(country, type, cases) %>%
group_by(country, type) %>%
summarise(total_cases = sum(cases)) %>%
pivot_wider(names_from = type,
values_from = total_cases) %>%
arrange(-confirmed)
#> # A tibble: 188 x 4
#> # Groups: country [188]
#> country confirmed death recovered
#> <chr> <int> <int> <int>
#> 1 India 60963 834 56110
#> 2 US 46808 1074 44205
#> 3 Colombia 12830 321 8943
#> 4 Argentina 7043 240 73147
#> 5 Mexico 6686 926 4118
#> 6 Russia 4892 130 6479
#> 7 Spain 3632 5 0
#> 8 Iraq 3396 67 2312
#> 9 Bangladesh 2996 33 1535
#> 10 Philippines 2900 18 273
#> 11 South Africa 2511 130 8925
#> 12 Iran 2345 184 1978
#> 13 Israel 1871 9 1082
#> 14 Bolivia 1693 49 930
#> 15 Indonesia 1693 59 1474
#> 16 Chile 1572 39 2199
#> 17 Saudi Arabia 1521 34 1640
#> 18 Romania 1215 35 274
#> 19 Ukraine 1211 29 590
#> 20 Turkey 1183 15 1185
#> 21 Venezuela 1138 9 2776
#> 22 Morocco 1132 17 861
#> 23 Panama 1070 16 1155
#> 24 Germany 1032 5 965
#> 25 Guatemala 979 11 853
#> 26 Ecuador 862 19 2
#> 27 Kazakhstan 691 211 975
#> 28 Japan 685 6 686
#> 29 Kuwait 668 4 731
#> 30 Nepal 638 4 171
#> 31 Costa Rica 636 11 148
#> 32 Dominican Republic 595 18 756
#> 33 Ethiopia 584 20 285
#> 34 Poland 551 12 273
#> 35 Honduras 531 9 156
#> 36 Pakistan 531 15 482
#> 37 Kenya 497 15 372
#> 38 Algeria 492 10 343
#> 39 Uzbekistan 443 4 712
#> 40 Nigeria 423 6 263
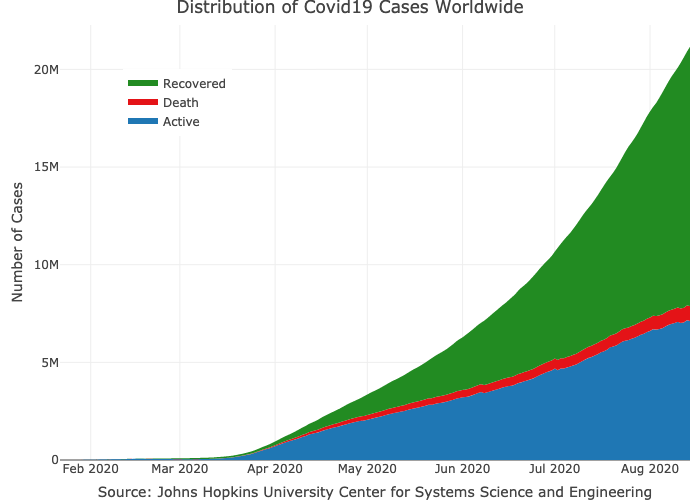
#> # … with 148 more rowsPlotting the total cases by type worldwide:
library(plotly)
coronavirus %>%
group_by(type, date) %>%
summarise(total_cases = sum(cases)) %>%
pivot_wider(names_from = type, values_from = total_cases) %>%
arrange(date) %>%
mutate(active = confirmed - death - recovered) %>%
mutate(active_total = cumsum(active),
recovered_total = cumsum(recovered),
death_total = cumsum(death)) %>%
plot_ly(x = ~ date,
y = ~ active_total,
name = 'Active',
fillcolor = '#1f77b4',
type = 'scatter',
mode = 'none',
stackgroup = 'one') %>%
add_trace(y = ~ death_total,
name = "Death",
fillcolor = '#E41317') %>%
add_trace(y = ~recovered_total,
name = 'Recovered',
fillcolor = 'forestgreen') %>%
layout(title = "Distribution of Covid19 Cases Worldwide",
legend = list(x = 0.1, y = 0.9),
yaxis = list(title = "Number of Cases"),
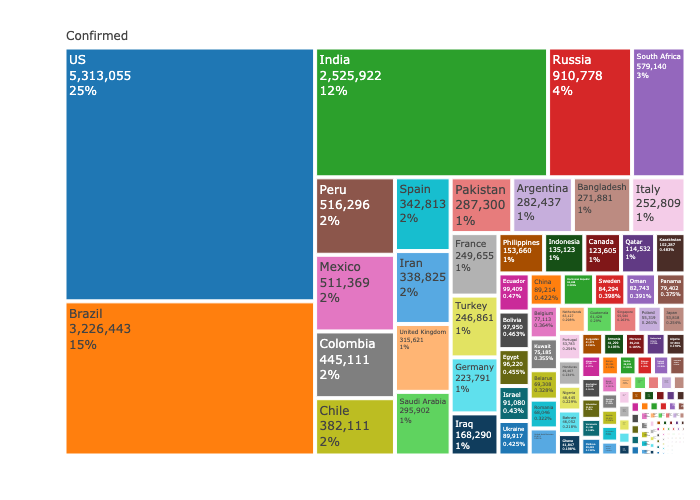
xaxis = list(title = "Source: Johns Hopkins University Center for Systems Science and Engineering"))Plot the confirmed cases distribution by counrty with treemap plot:
conf_df <- coronavirus %>%
filter(type == "confirmed") %>%
group_by(country) %>%
summarise(total_cases = sum(cases)) %>%
arrange(-total_cases) %>%
mutate(parents = "Confirmed") %>%
ungroup()
plot_ly(data = conf_df,
type= "treemap",
values = ~total_cases,
labels= ~ country,
parents= ~parents,
domain = list(column=0),
name = "Confirmed",
textinfo="label+value+percent parent")The raw data pulled and arranged by the Johns Hopkins University Center for Systems Science and Engineering (JHU CCSE) from the following resources:
- World Health Organization (WHO): https://www.who.int/
- DXY.cn. Pneumonia. 2020. http://3g.dxy.cn/newh5/view/pneumonia.
- BNO News:
https://bnonews.com/index.php/2020/02/the-latest-coronavirus-cases/
- National Health Commission of the People’s Republic of China (NHC):
http:://www.nhc.gov.cn/xcs/yqtb/list_gzbd.shtml - China CDC (CCDC):
http:://weekly.chinacdc.cn/news/TrackingtheEpidemic.htm
- Hong Kong Department of Health:
https://www.chp.gov.hk/en/features/102465.html
- Macau Government: https://www.ssm.gov.mo/portal/
- Taiwan CDC:
https://sites.google.com/cdc.gov.tw/2019ncov/taiwan?authuser=0
- US CDC: https://www.cdc.gov/coronavirus/2019-ncov/index.html
- Government of Canada:
https://www.canada.ca/en/public-health/services/diseases/coronavirus.html
- Australia Government Department of Health:
https://www.health.gov.au/news/coronavirus-update-at-a-glance
- European Centre for Disease Prevention and Control (ECDC): https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases
- Ministry of Health Singapore (MOH): https://www.moh.gov.sg/covid-19
- Italy Ministry of Health: http://www.salute.gov.it/nuovocoronavirus
- 1Point3Arces: https://coronavirus.1point3acres.com/en
- WorldoMeters: https://www.worldometers.info/coronavirus/
- COVID Tracking Project: https://covidtracking.com/data. (US Testing and Hospitalization Data. We use the maximum reported value from “Currently” and “Cumulative” Hospitalized for our hospitalization number reported for each state.)
- French Government: https://dashboard.covid19.data.gouv.fr/
- COVID Live (Australia): https://www.covidlive.com.au/
- Washington State Department of Health: https://www.doh.wa.gov/emergencies/coronavirus
- Maryland Department of Health: https://coronavirus.maryland.gov/
- New York State Department of Health: https://health.data.ny.gov/Health/New-York-State-Statewide-COVID-19-Testing/xdss-u53e/data
- NYC Department of Health and Mental Hygiene: https://www1.nyc.gov/site/doh/covid/covid-19-data.page and https://github.com/nychealth/coronavirus-data
- Florida Department of Health Dashboard: https://services1.arcgis.com/CY1LXxl9zlJeBuRZ/arcgis/rest/services/Florida_COVID19_Cases/FeatureServer/0 and https://fdoh.maps.arcgis.com/apps/opsdashboard/index.html#/8d0de33f260d444c852a615dc7837c86
- Palestine (West Bank and Gaza): https://corona.ps/details
- Israel: https://govextra.gov.il/ministry-of-health/corona/corona-virus/
- Colorado: https://covid19.colorado.gov/covid-19-data