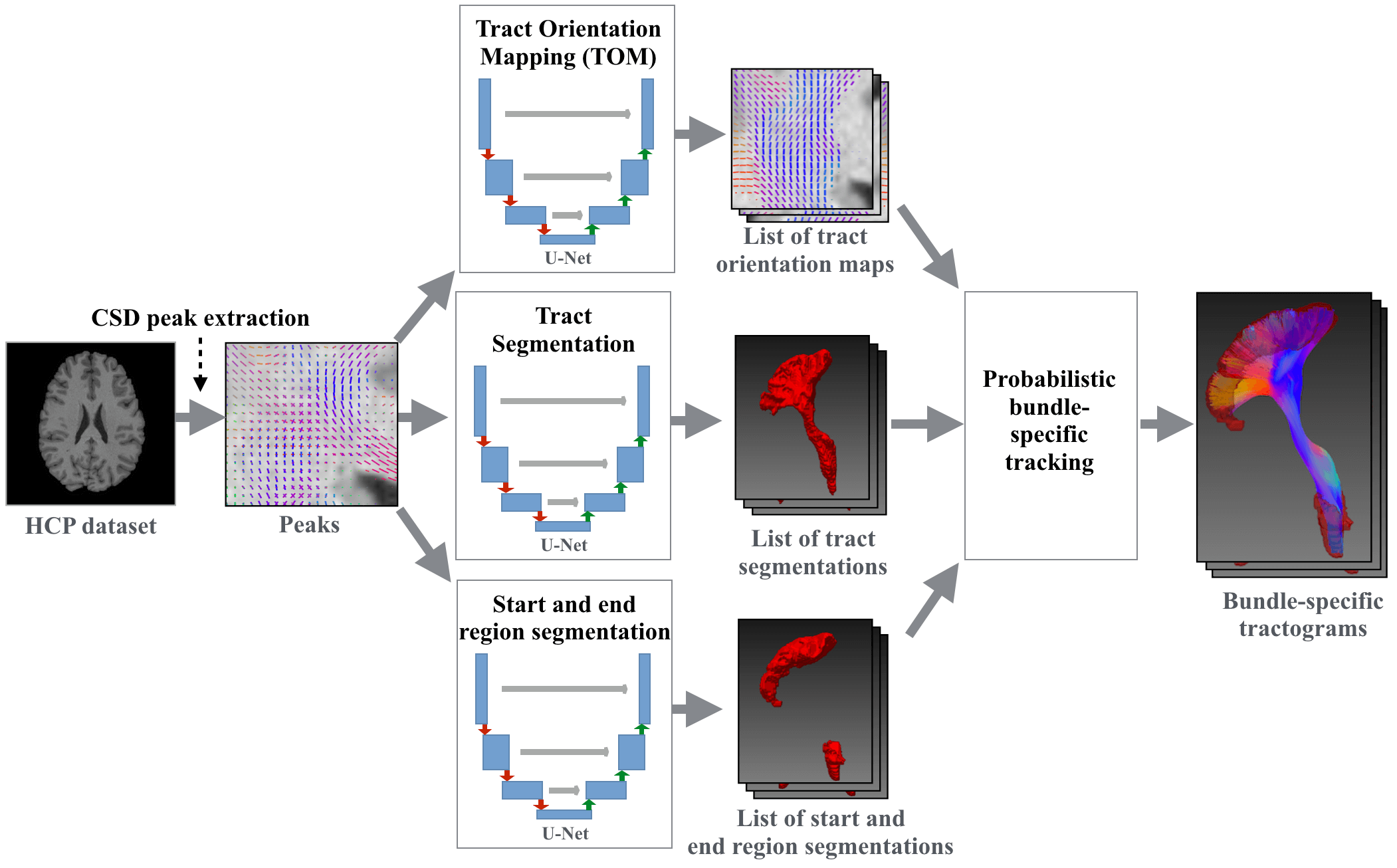
Tool for fast and accurate white matter bundle segmentation from Diffusion MRI. It can create bundle segmentations, segmentations of the endregions of bundles and Tract Orientation Maps (TOMs). Moreover, it can do tracking on the TOMs creating bundle-specific tractogram and do Tractometry analysis on those.
The tool works very well for data similar to the Human Connectome Project. For other MRI datasets it works well for all bundles except for the Commissure Anterior (CA) and the Fornix (FX) which are incomplete sometimes.
TractSeg is the code for the following papers. Please cite the papers if you use it.
- Tract Segmentation:
TractSeg - Fast and accurate white matter tract segmentation (free arxiv version) [NeuroImage 2018] - Tract Orientation Mapping (TOM):
Tract orientation mapping for bundle-specific tractography [MICCAI 2018] - Tracking on TOMs and Tractometry:
Combined tract segmentation and orientation mapping for bundle-specific tractography [Medical Image Analysis 2019]
TractSeg only runs on Linux and OSX. It works with Python 3.
- Pytorch
- Mrtrix 3
- FSL (if you already have a brain mask and do not use the
option
--preprocessthis is not needed)
Latest stable version:
pip install TractSeg
NOTE: See CHANGELOG for (breaking) changes of each version
You can also directly use TractSeg via Docker (contains all prerequisites).
sudo docker run -v /absolute/path/to/my/data/directory:/data \
-t wasserth/tractseg_container:master TractSeg -i /data/my_diffusion_file.nii.gz -o /data --raw_diffusion_input
On OSX you might have to increase the Docker memory limit from the default of 2GB to something like 7GB.
To segment the bundles on a Diffusion Nifti image run the following command. (Diffusion.bvals and Diffusion.bvecs have to be in the same directory
as the input image.)
(You can use the example image provided in this repository under examples.)
TractSeg -i Diffusion.nii.gz --raw_diffusion_input
This will create a folder tractseg_ouput inside of the same directory as your input file with one binary segmentation nifti image
for each bundle.
NOTE: Your input image should have the same orientation as MNI space. Moreover it should have isotropic spacing. See here for more details.
TractSeg -i my/path/my_diffusion_image.nii.gz
-o my/output/directory
--bvals my/other/path/my.bvals
--bvecs yet/another/path/my.bvec
--raw_diffusion_input
Use --help to see all options.
To avoid generating the MRtrix CSD peaks every time you run TractSeg you can also provide them directly by skipping the option --raw_diffusion_input`.
TractSeg -i my/path/my_mrtrix_csd_peaks.nii.gz
For each bundle create a Tract Orientation Map (Wasserthal et al., Tract orientation mapping for bundle-specific tractography). This gives you one peak per voxel telling you the main orientation of the respective bundle at this voxel. Can be used for bundle-specific tracking later on.
TractSeg -i peaks.nii.gz --output_type TOM
Peaks and streamlines can be visualized using for example MITK Diffusion.
NOTE: Peaks have to be flipped along the z-axis to be displayed correctly in MITK.
Get segmentations of the regions were the bundles start and end (helpful for filtering fibers that do not run from start until end).
TractSeg -i peaks.nii.gz --output_type endings_segmentation
Tracks on TOMs and only keeps fibers not leaving the bundle mask and starting and ending in the endpoint regions.
TractSeg -i peaks.nii.gz --output_type tract_segmentation
TractSeg -i peaks.nii.gz --output_type endings_segmentation
TractSeg -i peaks.nii.gz --output_type TOM
Tracking -i peaks.nii.gz
TractSeg also works with bedpostX as input. You have to pass dyads1.nii.gz as input and TractSeg will automatically
find all the other necessary bedpostX files (dyads2_thr0.05.nii.gz & dyads3_thr0.05.nii.gz. mean_f1-3samples
will be used for scaling the peaks.). This only works if you did not change the default bedpostX file naming.
TractSeg -i dyads1.nii.gz
Create map showing where the method is uncertain about its segmentation (uses monte carlo dropout: https://arxiv.org/abs/1506.02142)
TractSeg -i peaks.nii.gz --uncertainty
See Documentation of Tractometry.
Best pratices for standard usecases.
You can also check out the tutorial from OHBM 2019.
You can specify to only track a subset of bundles.
Tracking -i peaks.nii.gz --bundles CST_right,CA,IFO_right
The following list shows the index of each extracted bundle in the output file (if using --single_output_file).
0: AF_left (Arcuate fascicle)
1: AF_right
2: ATR_left (Anterior Thalamic Radiation)
3: ATR_right
4: CA (Commissure Anterior)
5: CC_1 (Rostrum)
6: CC_2 (Genu)
7: CC_3 (Rostral body (Premotor))
8: CC_4 (Anterior midbody (Primary Motor))
9: CC_5 (Posterior midbody (Primary Somatosensory))
10: CC_6 (Isthmus)
11: CC_7 (Splenium)
12: CG_left (Cingulum left)
13: CG_right
14: CST_left (Corticospinal tract)
15: CST_right
16: MLF_left (Middle longitudinal fascicle)
17: MLF_right
18: FPT_left (Fronto-pontine tract)
19: FPT_right
20: FX_left (Fornix)
21: FX_right
22: ICP_left (Inferior cerebellar peduncle)
23: ICP_right
24: IFO_left (Inferior occipito-frontal fascicle)
25: IFO_right
26: ILF_left (Inferior longitudinal fascicle)
27: ILF_right
28: MCP (Middle cerebellar peduncle)
29: OR_left (Optic radiation)
30: OR_right
31: POPT_left (Parieto‐occipital pontine)
32: POPT_right
33: SCP_left (Superior cerebellar peduncle)
34: SCP_right
35: SLF_I_left (Superior longitudinal fascicle I)
36: SLF_I_right
37: SLF_II_left (Superior longitudinal fascicle II)
38: SLF_II_right
39: SLF_III_left (Superior longitudinal fascicle III)
40: SLF_III_right
41: STR_left (Superior Thalamic Radiation)
42: STR_right
43: UF_left (Uncinate fascicle)
44: UF_right
45: CC (Corpus Callosum - all)
46: T_PREF_left (Thalamo-prefrontal)
47: T_PREF_right
48: T_PREM_left (Thalamo-premotor)
49: T_PREM_right
50: T_PREC_left (Thalamo-precentral)
51: T_PREC_right
52: T_POSTC_left (Thalamo-postcentral)
53: T_POSTC_right
54: T_PAR_left (Thalamo-parietal)
55: T_PAR_right
56: T_OCC_left (Thalamo-occipital)
57: T_OCC_right
58: ST_FO_left (Striato-fronto-orbital)
59: ST_FO_right
60: ST_PREF_left (Striato-prefrontal)
61: ST_PREF_right
62: ST_PREM_left (Striato-premotor)
63: ST_PREM_right
64: ST_PREC_left (Striato-precentral)
65: ST_PREC_right
66: ST_POSTC_left (Striato-postcentral)
67: ST_POSTC_right
68: ST_PAR_left (Striato-parietal)
69: ST_PAR_right
70: ST_OCC_left (Striato-occipital)
71: ST_OCC_right
TractSeg was also trained on the bundles provided by XTRACT. These bundles are slightly differently defined. They tend to be more specific but therefore also a bit less complete. Depending on your application this might be of interest for you. Using TractSeg instead of XTRACT has the advantage of being at least 10x faster. Please cite XTRACT if you use this.
NOTE: This is only supported for output type
tract_segmentationanddm_regression.
TractSeg -i peaks.nii.gz --tract_definition xtract
import nibabel as nib
import numpy as np
from tractseg.python_api import run_tractseg
peaks = nib.load("tests/reference_files/peaks.nii.gz").get_data()
segmentation = run_tractseg(peaks)You can use different types of tracking:
-
"Probabilistic" tracking on TOM peaks [default].
Tracking -i peaks.nii.gz
Probabilistic means that at each step a small random factor will be added to the direction given by the TOM peaks. If not doing this on low resolution data it sometimes gets difficult finding fibers running from start to end and covering the whole bundle. -
Probabilistic tracking on original FODs.
Tracking -i WM_FODs.nii.gz --track_FODs iFOD2
Is calling Mrtrix iFOD2 tracking internally. Does not use TOM peaks but the original FODs. The results will get filtered by the bundle mask and have to start and end in the endings masks.
You can use the option --tracking_format to define the file format of the streamline files.
The default is trk_legacy. The resulting streamlines are displayed correctly in MITK Diffusion.
If you are using TrackVis you should use the format trk.
NOTE: When calling
Tractometryyou have to set the same tracking format as was used inTracking.
The input image must have the same "orientation" as the Human Connectome Project data (MNI space)
(LEFT must be on the same side as LEFT of the HCP data) and have isotropic spacing.
If the image orientation and the gradient orientation of your data is the same as in examples/Diffusion.nii.gz
you are fine. Otherwise you should use fslreorient2std or rigidly register your image to MNI space (the brains
do not have to be perfectly aligned but must have the same LEFT/RIGHT orientation).
You can use the following FSL commands to rigidly register you image to MNI space (uses
the FA to calculate the transformation as this is more stable):
calc_FA -i Diffusion.nii.gz -o FA.nii.gz --bvals Diffusion.bvals --bvecs Diffusion.bvecs \
--brain_mask nodif_brain_mask.nii.gz
flirt -ref tractseg/tractseg/resources/MNI_FA_template.nii.gz -in FA.nii.gz \
-out FA_MNI.nii.gz -omat FA_2_MNI.mat -dof 6 -cost mutualinfo -searchcost mutualinfo
flirt -ref tractseg/tractseg/resources/MNI_FA_template.nii.gz -in Diffusion.nii.gz \
-out Diffusion_MNI.nii.gz -applyxfm -init FA_2_MNI.mat -dof 6
cp Diffusion.bvals Diffusion_MNI.bvals
rotate_bvecs -i Diffusion.bvecs -t FA_2_MNI.mat -o Diffusion_MNI.bvecsTo enforce isotropic spacing you can replace -applyxfm by -applyisoxfm <your_spacing>.
To move the results back to subject space you can use the following commands:
convert_xfm -omat MNI_2_FA.mat -inverse FA_2_MNI.mat # invert transformation
flirt -ref FA.nii.gz -in my_bundle.nii.gz -out my_bundle_subject_space.nii.gz \
-applyxfm -init MNI_2_FA.mat -dof 6 -interp trilinear # use -interp nearestneighbour for TOM
# Do not run this line for TOM
fslmaths my_bundle_subject_space.nii.gz -thr 0.5 -bin my_bundle_subject_space.nii.gz # float to binary The option --preprocess will automatically rigidly register the input image to MNI space, run TractSeg and then
convert the output back to subject space. However, this does not work if you are using the option
--csd_type csd_msmt_5tt, because the T1 image will not automatically be registered to MNI space. --preprocess can
also not be used if you want Tract Orientation Maps (TOMs). In this case you will have to run the registration commands
stated above manually.
Make sure your input image is in MNI space. Even if the input image is in MNI space the Mrtrix peaks might still be flipped.
In those cases you should view the peaks in mrview and make sure they have the proper
orientation. Otherwise you might have to flip the sign along the x, y or z axis using the following command:
flip_peaks -i my_peaks.nii.gz -o my_peaks_flip_y.nii.gz -a y
If the spacing is only slightly anisotropic (e.g. 1.9mm x 1.9mm x 2mm) it still works fine. If the spacing is heavily anisotropic (e.g. 1mm x 1mm x 2mm) results will be a lot better if you resample your image to isotropic spacing first.
You can use the following options to improve your results:
--super_resolution The input image is upsampled to 1.25mm resolution (the resolution TractSeg was trained on) and
finally downsampled back to the original resolution. Using --super_resolution will output the image at 1.25mm.
Especially if image resolution is low parts of the CA can get lost during downsampling.
Per default the pretrained weights will be downloaded to and loaded from ~/.tractseg/. You can change this directory
by adding weights_dir=/absolute/path_to_where/you_want_it to ~/.tractseg/config.txt in a new line (if the file does
not exist yet you have to create it).
Normally the pretrained weights will automatically be downloaded in the background right when they are needed. In some
cases you might want to download all of them at once. To do so you can simply run download_all_pretrained_weights and
the weights will be download to ~/.tractseg/ or the location you specified in ~/.tractseg/config.txt.
You can check if you installed Mrtrix correctly if you can run the following command on your terminal:
dwi2response -help
You can check if you installed FSL correctly if you can run the following command on your terminal:
bet -help
TractSeg uses these commands so they have to be available.
pip install https://github.com/MIC-DKFZ/TractSeg/archive/master.zip
TractSeg uses a pretrained model. You can also train your own model on your own data, however TractSeg is not optimised to make this convenient, as most people will use the pretrained model. The following guide is quite short and you might have problems following every step. Contact the author if you need help training your own model.
- Use your own data or download the data from Zenodo
- If you have streamlines you have to transform them to binary masks.
You can use this
script for that.
(If you want to train a TOM model you have to create peak maps from the streamlines. This can be done
by using the MITK Diffusion miniapp
MitkFiberDirectionExtraction. See the following two issues for more details: 82, 92) - Install TractSeg from local source code:
git clone https://github.com/MIC-DKFZ/TractSeg.git pip install -e TractSeg - Install BatchGenerators:
git clone https://github.com/MIC-DKFZ/batchgenerators.git pip intall -e batchgenerators - The folder structure of your training data should be the following:
custom_path/HCP/subject_01/ '-> mrtrix_peaks.nii.gz (mrtrix CSD peaks; shape: [x,y,z,9]) '-> bundle_masks.nii.gz (Reference bundle masks; shape: [x,y,z,nr_bundles]) custom_path/HCP/subject_02/ ... - Preprocess the data using
tractseg/data/preprocessing.pyto remove all non-brain area (crop to brain bounding box). Adapt the data pathes intractseg/data/preprocessing.pyto fit your data (look for#todo: adaptinside of the file.) - Adapt the file
tractseg/config/custom/My_custom_experiment.py. - Create a file
~/.tractseg/config.txt. This contains the path to your data directoryworking_dir=XXX, e.g.working_dir=custom_path. - Adapt
tractseg.data.dataset_specific_utils.get_bundle_names()with the bundles you use in your reference data. - Adapt
tractseg.data.dataset_specific_utils.get_labels_filename()with the names of your label files. - Adapt
tractseg.data.Subjectswith the list of your subject IDs. - Run
ExpRunner --config My_custom_experiment custom_path/hcp_exp/My_custom_experimentcontains the results
To build a docker container with all dependencies run the following command in project root:
sudo docker build -t tractseg_container -f Dockerfile_CPU .
Copyright © German Cancer Research Center (DKFZ), Division of Medical Image Computing (MIC). Please make sure that your usage of this code is in compliance with the code license.