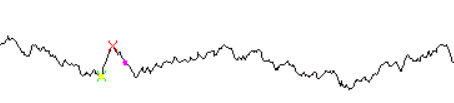
Electrophysiological recordings are time series data. Our data is a result of recording skeletal muscle membrane potential, which consists of noise and miniature end plate potentials (marked in the picture below).
Currently the data is processed manually, what takes hours of work. Automatization is urgent. Some existing methods (for example, function find_peaks in library scipy.signal) are not able to solve this problem, as they cannot discriminate miniature end plate potentials from noise.
At the same time neural networks are widely used for time series processing. Thus, 1-D convolution neural networks are used, for example, for EEG and ECG processing (Kiranyaz et al., 2019, Zahid et al., 2020). Autoencoders are also used for anomaly detection in time series (Wei et al., 2022, Timeseries anomaly detection using an Autoencoder).
The aim of this project is to create a tool for automatic electrophysiological data processing.
Tasks:
- Preprocess data.
- Develop 1D convolution NN architecture based on article Zahid et al., 2020 (segmentation task).
- Develop Autoencoder architecture based on example Timeseries anomaly detection using an Autoencoder (anomaly detection task).
Recordings of skeletal muscle membrane potential are time series. Each recording lasts approximately 60 sec, sampling frequency is 10.000/sec. This work includes 133 recordings (1.9 GB). You can find these recordings in this repository in folder data.
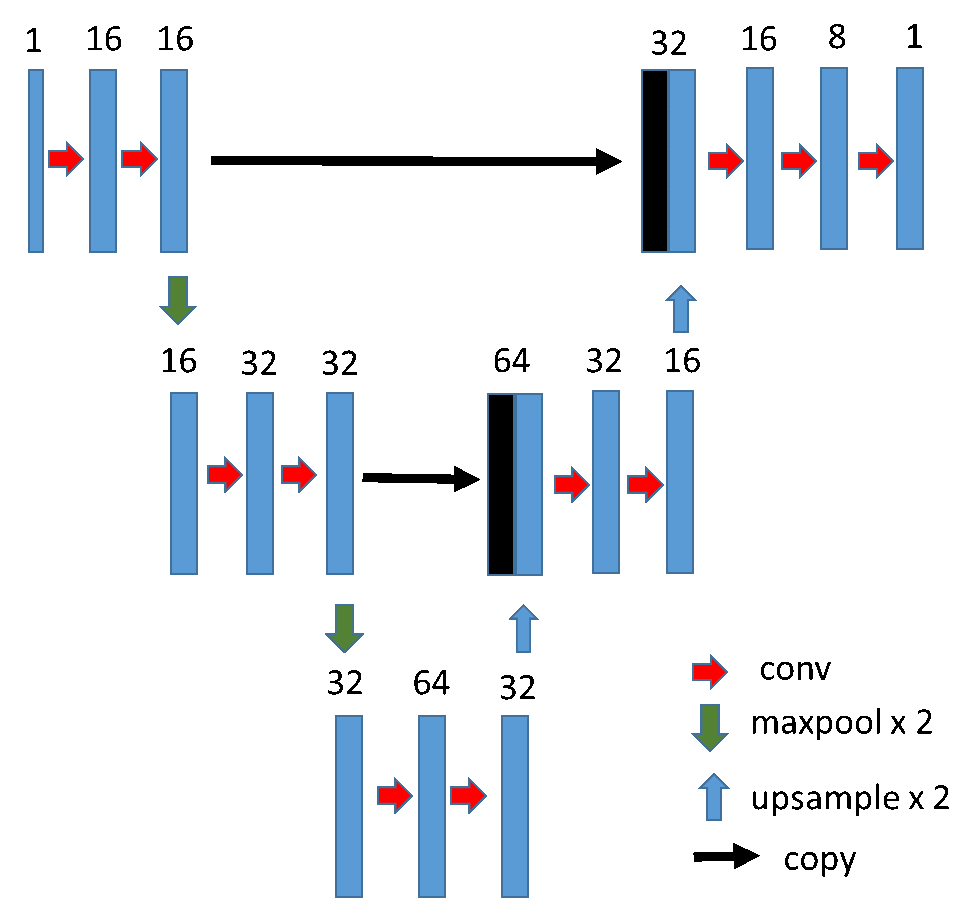
During this project two NN architectures were developed: 1D MiniUnet and Autoencoder. 1D MiniUnet seems to be a more successful architecture. Intersection over Union was 0.91 for patches and about 0.5 for whole recordings. 1D MiniUnet can successfully detect the borders of miniature end plate potentials. You can see the 1D MiniUnet architecture below.
Autoencoder can also discriminate miniature end plate potentials, but it also detects other anomalies. Moreover, it requires "normal" data for training (noise without miniature end plate potentials). We lack recordings without miniature end plate potentials in our experiments. While training Autoencoder on data with them gives worse results. Comparison of two models (autoencoder trained in two different ways) is presented in table below.
| Model | Precision | Recall | F1 score |
|---|---|---|---|
| 1D MiniUnet | 0.73 | 1 | 0.85 |
| Autoencoder, trained on normal data | 0.82 | 0.64 | 0.72 |
| Autoencoder, trained on anomalous data | 0.52 | 0.58 | 0.55 |
In this repository you can see several notebooks and a folder with files ("data"). You do not need notebook "File_markup.ipynb" because files are already marked up. You can see notebook 1D MiniUnet, notebook Autoencoder, trained on normal data, and Autoencoder, trained on anomalous data. You can launch the code in that order, as it is in these notebooks. There are comments on what everys step does.
Launch these notebooks in google colab.
The project was done with Python 3.8.5.
-
Kiranyaz S., Ince T., Abdeljaber O., Avci O., Gabbouj M. 1-D Convolutional Neural Networks for Signal Processing Applications // ICASSP, IEEE Int. Conf. Acoust. Speech Signal Process. - Proc. 2019. Т. 2019- May. С. 8360–8364.
-
Lun X., Yu Z., Chen T., Wang F., Hou Y. A Simplified CNN Classification Method for MI-EEG via the Electrode Pairs Signals // Front. Hum. Neurosci. 2020. Т. 14. С. 338.
-
Pol A. A., Azzolini V., Cerminara G., Guio F. De, Franzoni G., Pierini M., Siroký F., Vlimant J.-R. Anomaly detection using Deep Autoencoders for the assessment of the quality of the data acquired by the CMS experiment // EPJ Web Conf. 2019. Т. 214. С. 06008.
-
Wei Y., Jang-Jaccard J., Xu W., Sabrina F., Camtepe S., Boulic M. LSTM-Autoencoder based Anomaly Detection for Indoor Air Quality Time Series Data // 2022.
-
Wu M., Lu Y., Yang W., Wong S. Y. A Study on Arrhythmia via ECG Signal Classification Using the Convolutional Neural Network // Front. Comput. Neurosci. 2021. Т. 14. С. 106.
-
Zahid M. U., Kiranyaz S., Ince T., Devecioglu O. C., Chowdhury M. E. H., Khandakar A., Tahir A., Gabbouj M. Robust R-Peak Detection in Low-Quality Holter ECGs using 1D Convolutional Neural Network // IEEE Trans. Biomed. Eng. 2020. Т. 69. № 1. С. 119–128.
-
https://keras.io/examples/timeseries/timeseries_anomaly_detection/
-
https://stepik.org/lesson/532600/step/1?auth=login&unit=717336