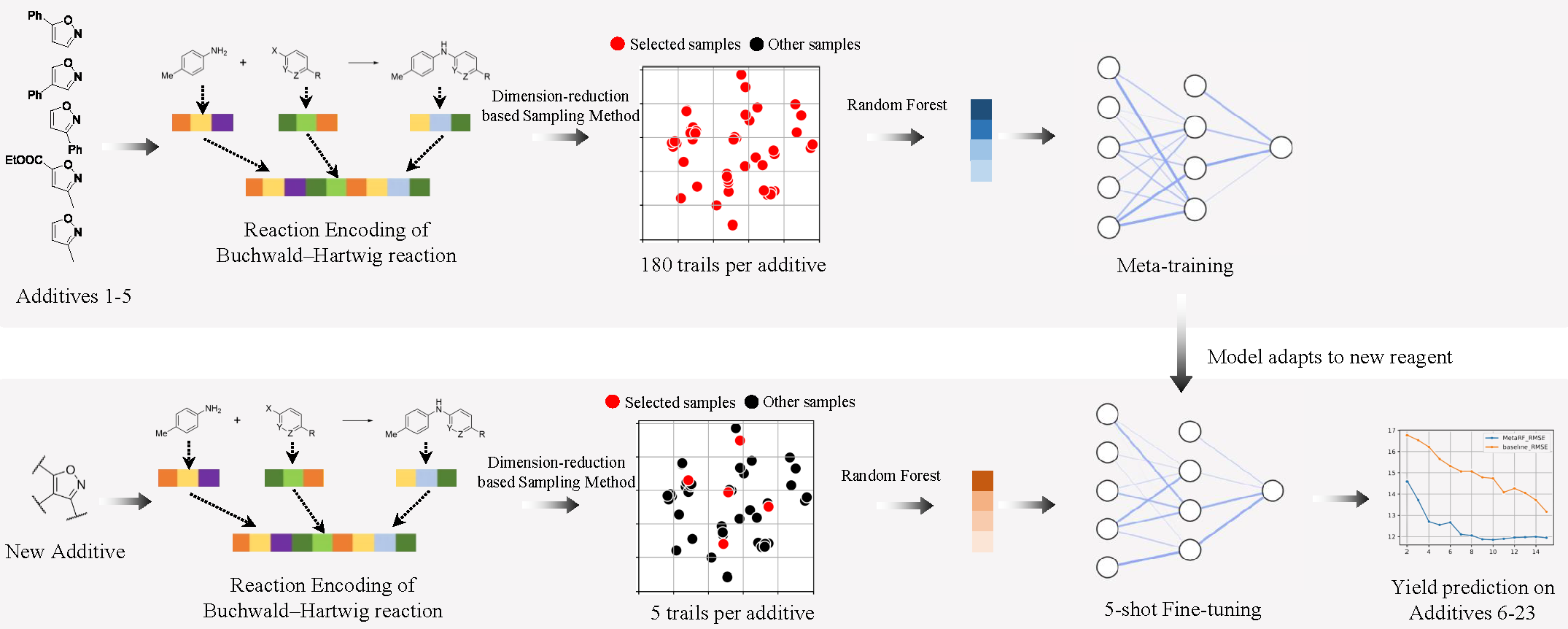
This is the implementation for our paper "MetaRF: Differentiable Random Forest for Reaction Yield Prediction with a Few Trails".
- Check dependencies
- tensorflow==2.9.2
- kennard-stone==1.1.2
- numpy
- pandas
- sklearn
- Clone this repo
git clone https://github.com/Nikki0526/MetaRF.git
- Run
$ data_preprocessing.pyto preprocess the data. - This step includes random forest module and dimension-reduction module.
- The original reaction and yield data in this paper is from [1], [2] and [3].
- We also provide the data after preprocessing in
/data(Two datasets are too large for github and they can be downloaded from Google Drive).
- Run
$ train.pyto perform meta-training and model saving. - Our trained model can be downloaded from Google Drive.
- Run
$ test.pyto perform few-shot fine-tuning, dimension-reduction based sampling method and model evaluation. - We use relative path in this repository. Please place the downloaded model in the
/modelfolder. - [update] We add more baseline comparision in
$ baseline.py, including RXNFP [4], DRFP [5], etc.
We provide a step-by-step tutorial that includes the whole workflow (including Data preprocessing, Model training, Model fine-tuning and testing, Baseline comparision) in $ Workflow of MetaRF - Tutorial.ipynb. We also provide a colab version, which can help users easily access our code and environment by clicking:
Note:
In this tutorial, we take the procedure for Buchwald Hartwig HTE dataset as an example. The other two datasets share the same procedure.
For further question about the code, please contact 'kexinchen0526@gmail.com'.
[1] Ahneman, D.T., Estrada, J.G., Lin, S., Dreher, S.D., Doyle, A.G.: Predicting reaction performance in c–n cross-coupling using machine learning. Science 360(6385), 186–190(2018).
[2] Perera, D., Tucker, J.W., Brahmbhatt, S., Helal, C.J., Chong, A., Farrell, W., Richardson, P., Sach, N.W.: A platform for automated nanomole-scale reaction screening and micromole-scale synthesis in flow. Science 359(6374), 429–434(2018).
[3] Saebi, M., Nan, B., Herr, J., Wahlers, J., Guo, Z., Zura ́nski, A., Kogej, T., Norrby, P.-O., Doyle, A., Wiest, O., et al.: On the use of real-world datasets for reaction yield prediction. ChemRxiv (2021).
[4] Schwaller, P., Vaucher, A. C., Laino, T., & Reymond, J. L. (2021). Prediction of chemical reaction yields using deep learning. Machine learning: science and technology, 2(1), 015016.
[5] Probst, D., Schwaller, P., & Reymond, J. L. (2022). Reaction classification and yield prediction using the differential reaction fingerprint DRFP. Digital discovery, 1(2), 91-97.