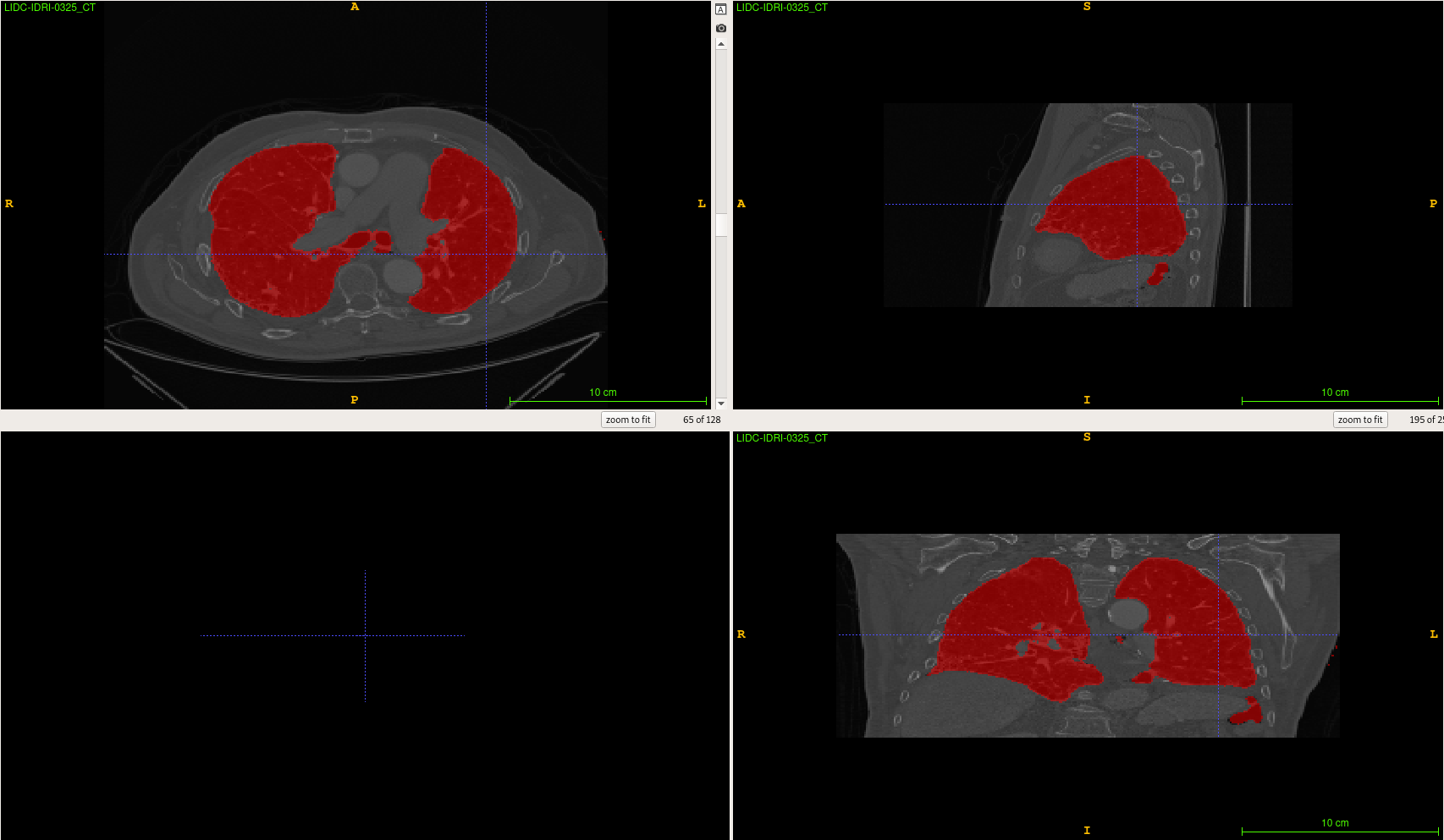
Lung Segmentation
Lung Segmentation using a U-Net model on 3D CT scans.
Current results example :
Getting started
Installation
Our base wmlce conda environment does not come with SimpleITK, pynrrd and pysftp (for MLFlow integration), two required python libraries to run this code.
- To install
pynrrd:
$ pip install pynrrd
- To install
mlflow:
$ pip install mlflow
- To install
pysftp(for MLflow integration):
$ sudo apt install libffi-dev
$ pip install pysftp==0.2.8
-
You also need to add your MLFlow sftp host to
~/.ssh/known-hosts -
To install
SimpleITK(from wheel):- Python
3.6.x:
- Python
$ pip install /wmlce/data/install-files/SimpleITK-1.2.0+gd6026-cp36-cp36m-linux_ppc64le.whl
- Python
3.7.x:
$ pip install /wmlce/data/install-files/SimpleITK-1.2.0+gd6026-cp37-cp37m-linux_ppc64le.whl
- If you do not have access to the
whlfile, you need to build it (on power pc):
$ cd /wmlce/data/install-files
$ wget https://github.com/SimpleITK/SimpleITK/releases/download/v1.2.0/SimpleITK-1.2.0.zip
$ unzip SimpleITK-1.2.0.zip
$ mkdir SimpleITK-build/ && cd SimpleITK-build/
$ cmake ../SimpleITK-1.2.0/SuperBuild/
$ make -j100 # extra long
$ cd SimpleITK-build/Wrapping/Python
$ python Packaging/setup.py bdist_wheel
Tree
.
+-- data/
+-- dataset.py : Class describing the dataset we use for lung segmentation
+-- utils.py : Script for manipulating medical files
+-- eval.py
+-- model : Pre-trained pytorch model
+-- model.py : U-Net model definition
+-- predict.py : Inference script to run infer lung mask on a CT-scan
+-- README.md : This documentation file
+-- train.py : Train script to train a new lung segmentation model
Data
The data used is the TCIA LIDC-IDRI dataset Standardized representation (download here), combined with matching lung masks from LUNA16 (not all CT-scans have their lung masks in LUNA16 so we need the list of segmented ones).
3 parameters have to be fulfilled to use available data:
labelled-list: path to thepicklefile containing the list of CT-scans from the TCIA LIDC-IDRI dataset for which we have access to the lung segmentation masks through the LUNA16 dataset.scans: path to the TCIA LIDC-IDRI dataset.masks: path to the LUNA16 dataset containing lung masks.
You can manipulate data trough the data/dataset.py (class describing our lung segmentation dataset) and data/utils.py (tools for manipulating medical files) files.
Predictions
To perform predictions on unseen CT-scans, run for example (wmlce on powerai):
$ data=/wmlce/data/medical-datasets/LIDC-IDRI/LIDC-IDRI-0325/1.3.6.1.4.1.14519.5.2.1.6279.6001.815399168774050638734383723372/1.3.6.1.4.1.14519.5.2.1.6279.6001.725023183844147505748475581290/LIDC-IDRI-0325_CT.nrrd
$ output_path=/wmlce/data/projects/lung_segmentation/output/preds
$ nb_classes=1
$ start_filters=32
$ model=/wmlce/data/projects/lung_segmentation/model
$ python3 predict.py -d $data -o $output_path -m $model -c $nb_classes -f $start_filters -t [-e]
- See
python3 predict.py --helpfor more information.
Evaluation
To perform evaluation using the existing model, run for example (wmlce on powerai):
$ LABELLED_LIST=/wmlce/data/medical-datasets/labelled.pickle
$ MASKS=/wmlce/data/medical-datasets/lung_masks_LUNA16
$ SCANS=/wmlce/data/medical-datasets/LIDC-IDRI
$ NB_CLASSES=1
$ START_FILTERS=32
$ python3 eval.py --labelled-list $LABELLED_LIST --masks $MASKS --scans $SCANS --nb-classes $NB_CLASSES --start-filters $START_FILTERS
- See
python3 eval.py --helpfor more information.
Training
To run training:
python data/preprocessing.py -s /wmlce/data/medical-datasets/LUNA16/raw/ -l /wmlce/data/medical-datasets/LUNA16/seg-lungs-LUNA16/ -o output/preprocessing/ -v
python train.py -d output/preprocessing/
- See
python train.py --helpfor more information