The smms package allows you to fit Semi-Markovian multi-state models
to panel datasets. The package constructs and optimises the likelihood
of arbitrary multi-state models where the possible state transitions can
be described by an acyclic graph with one or more initial states and one
or more absorbing states.
Designed for data where the exact state transitions are not necessarily observed, so that we have interval censoring of the transition times. Data like these are sometimes referred to as panel data.
The methodology is explained in the paper A new framework for
semi-Markovian parametric multi-state models with interval
censoring
by Aastveit, Cunen and Hjort (2022). The code used for the application
and simulations in the paper is available in the scripts folder.
In order to directly install the package from github, you need the
package devtools.
install.packages("devtools")
devtools::install_github("NorskRegnesentral/smms")The main function in this package is the smms function, which is used
to fit a multi-state model. The user needs to provide a datasets of
states and time-points for multiple units of observation, typically
referred to as patients, a graph describing the states and the
possible transitions between them, and a set of parametric densities,
and survival functions.
In the next section, we will illustrate the use of the smms function
in a simple example.
We will use the CAV dataset from the msm package (Jackson 2011) as an
illustration. The dataset monitors a number of patients for a number of
years after heart transplantation. Coronary allograft vasculopathy (CAV)
is a condition potentially occurring after hear transplantation. At each
time-point the patients are assigned to one of four states: well, mild
CAV, severe CAV and death. The time of death is recorded precisely, but
the times of entrance into the CAV-states are interval censored
Here we see the observations belonging to two patients. Note here that the states were originally numbered from 1 to 4, but for the sake of this illustration I have changed the state names to “well”, “mild”, “severe” and “death”. This is to demonstrate that the package accepts names as both numbers and strings. After deleting some observations that are deemed incorrect (because they appear to get better, see next paragraph), we end up with 2398 observations in 556 different patients.
library(smms)
library(igraph) # For specifying the multi-state graph
library(msm) # To get the CAV dataset
dd = cav
dd = dd[!is.na(dd$pdiag),]
# Remove observations where the patient appears to go back to a previous state (assumed to be impossible):
id_wrong = unique(dd$PTNUM[which(dd$state!=dd$statemax)])
dd = dd[-which(dd$PTNUM %in% id_wrong),]
# Change state names from 1,2,3,4 to well, mild, severe, death
tab = data.frame(state=1:4,name=c("well","mild","severe","death"))
dd$state = tab$name[match(dd$state,tab$state)]
dd = dd[ ,-c(2, 5, 7, 9, 10)]
colnames(dd)[1:2] <- c("patient","time") # rename relevant columns (necessary in current version)
print(dd[1:11,])## patient time dage pdiag state
## 1 100002 0.000000 21 IHD well
## 2 100002 1.002740 21 IHD well
## 3 100002 2.002740 21 IHD mild
## 4 100002 3.093151 21 IHD mild
## 5 100002 4.000000 21 IHD mild
## 6 100002 4.997260 21 IHD severe
## 7 100002 5.854795 21 IHD death
## 8 100003 0.000000 17 IHD well
## 9 100003 1.189041 17 IHD well
## 10 100003 2.008219 17 IHD severe
## 11 100003 2.991781 17 IHD death
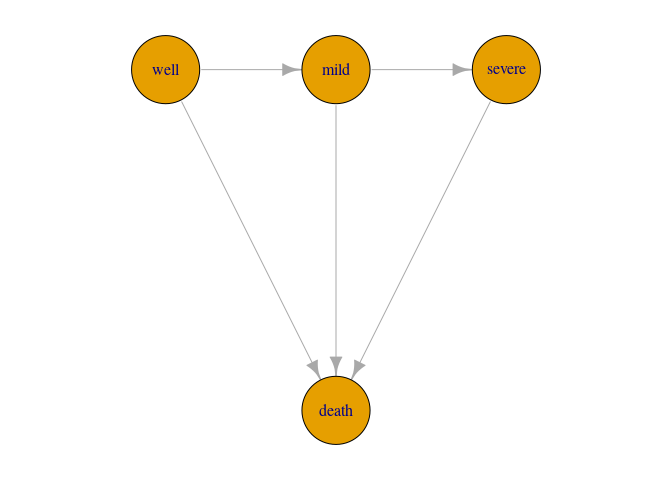
Here we assume a four-state illness death model, since we consider CAV to be irreversible (so we do not allow for patients to move back to less severe states). It is convenient to stick to the same state names as in the dataset when specifying the model graph.
# Specify the graph:
gg = graph_from_literal("well"--+"mild"--+"severe"--+"death", "well"--+"death", "mild"--+"death")
par(mar=c(1,1,1,1))
plot(gg,layout=layout_with_sugiyama(gg,layers=c(1,1,1,2))$layout,vertex.size=40)Then, the user has to specify parametric models for all transition times
(meaning one for each edge in the graph). In the current version of the
package, these models have to be specified by providing density
functions (in a specific format detailed below), as well as the
corresponding survival functions. The functions will look like the
following if one chooses to use simple exponential models for all
transitions (i.e. meaning that we are fitting a homogeneous Markov model
which could have been fitted with the msmpackage too - but this is for
the sake of a simple illustration):
f_01 = function(param, x, tt){dexp(tt,exp(param[1]))}
f_12 = function(param, x, tt){dexp(tt,exp(param[2]))}
f_23 = function(param, x, tt){dexp(tt,exp(param[3]))}
f_03 = function(param, x, tt){dexp(tt,exp(param[4]))}
f_13 = function(param, x, tt){dexp(tt,exp(param[5]))}
S_01 = function(param, x, tt){1-pexp(tt,exp(param[1]))}
S_12 = function(param, x, tt){1-pexp(tt,exp(param[2]))}
S_23 = function(param, x, tt){1-pexp(tt,exp(param[3]))}
S_03 = function(param, x, tt){1-pexp(tt,exp(param[4]))}
S_13 = function(param, x, tt){1-pexp(tt,exp(param[5]))}Important to note:
- the names of the functions: these have to be in the form
f_ijandS_ijwith i indicating the source state (in the internal numbering system) and j indicating the receiving state. More details on the naming convention below. - the arguments of the functions: these should always be given as
(param, x, tt)as above.xwill point to the vector of measured covariates (for a patient) when these are present. When there are no covariates, like in this example,xshould still be present as an argument, but will not be called within the functions. In the vignettes we include examples with covariates. - the scale of the parameters: for the sake of stable optimisation
it is convinient that the parameters live on the real line (instead
of the positive half-line as in the common parameterisation of the
exponential distribution). Therefore, we include an exponential
transformation of the
paramvector, and we recommend that transformation for all positive parameters. - the ordering of the parameters:
paramdenotes the full parameter vector for the model. - Survival functions should be written so that they return 1 when
ttis negative. Using build-in R CDFs for distributions over the positive half-line will ensure this. Otherwise, if the user codes the survival functions herself, she should ensure that they return 1 whenttis negative. We include an example with user-specified distribution functions in the vignettes.
As we saw above, the user has to follow a strict naming convention when
specifying the densities and survival functions: within the package, the
states are numbered from 0 to k-1 (k being the number of states), in a
specific order which depends on the graph. To find out how the user
defined state names relate to the internal numbering system, use the
names_of_survival_density function. This allways recommended before
specifying the model:
print(names_of_survival_density(gg))## edge_name survival_name density_name from_prev to_prev type
## 1 01 S_01 f_01 well mild trans
## 2 03 S_03 f_03 well death abs
## 3 12 S_12 f_12 mild severe trans
## 4 13 S_13 f_13 mild death abs
## 5 23 S_23 f_23 severe death abs
Here we see for example that the density for the edge between “mild” and
“severe” should be named f_12(as we do above).
Now we have everything in place in order to fit the multi-state model we have specified above:
startval <- c(-2.5,-1.1,-1.2,-3.1,-2.8)
mlo <- smms(startval,dd,gg, mc_cores = 1, hessian_matrix = T)One needs some start values for the optimisation, and we will soon add a function which calculates good starting values for a given model. Increasing the number of cores will make optimisation faster (but will not work on Windows machines). Here we choose to compute the hessian matrix too, which takes a bit more time. With one core this might take something like 5 minutes to compute on an ordinary laptop.
We can compute AIC, and look at the estimated parameters and approximate 95% confidence intervals.
# Compute AIC (higher values are better with this definition)
aic <- (-2*mlo$opt$objective)-2*length(mlo$opt$par) #-2887.1
# Look at estimates and 95% confidence intervals.
# On the -Inf to Inf scale:
print(round(est_ci(mlo$opt$par,mlo$hess),2)) ## estimate lower.ci upper.ci
## 1 -2.51 -2.66 -2.36
## 2 -1.11 -1.36 -0.86
## 3 -1.24 -1.48 -1.00
## 4 -3.11 -3.33 -2.90
## 5 -2.76 -3.67 -1.84
# On the 0 to Inf scale (on the transition intensity scale):
round(exp(est_ci(mlo$opt$par,mlo$hess)),2) ## estimate lower.ci upper.ci
## 1 0.08 0.07 0.09
## 2 0.33 0.26 0.42
## 3 0.29 0.23 0.37
## 4 0.04 0.04 0.06
## 5 0.06 0.03 0.16
- Jackson CH (2011). Multi-State Models for Panel Data: The msm Package for R. Journal of Statistical Software, 38, 1–29.