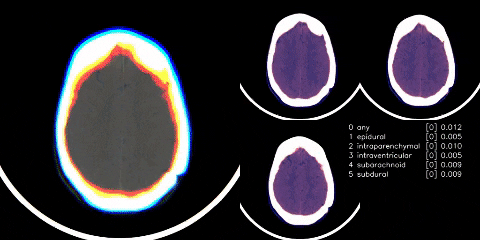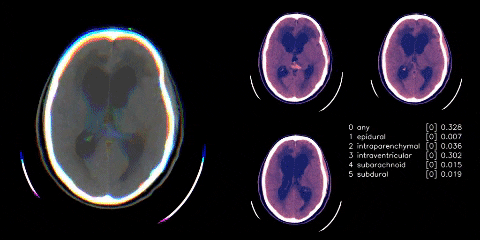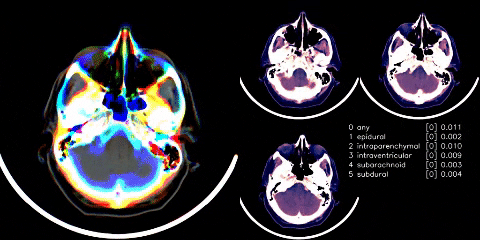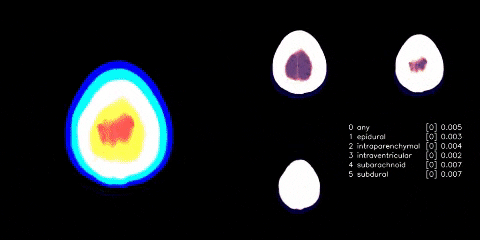
Hosted on Kaggle
Sponsored by RSNA
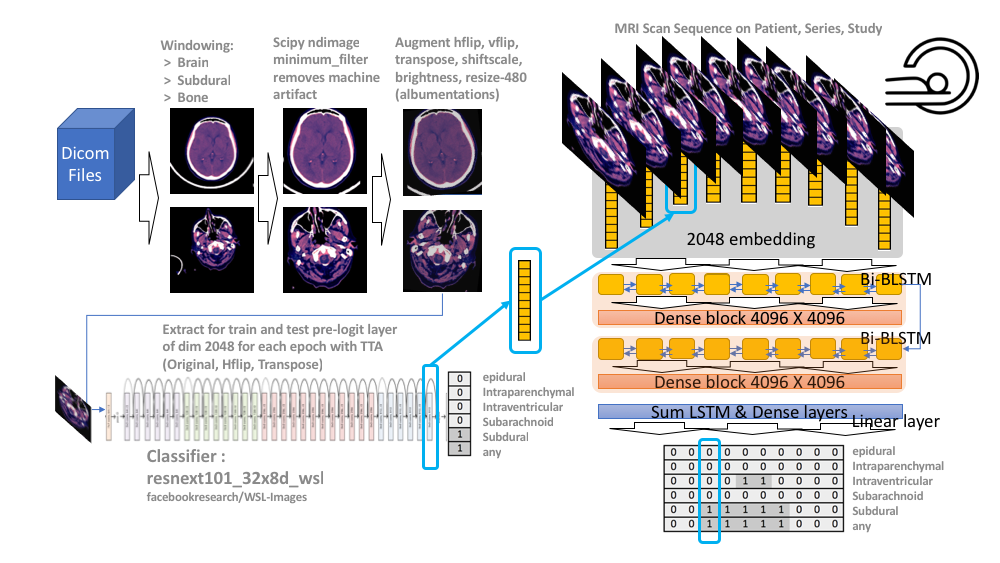
In general we just have a single image classifier, data split on 5 folds, we only trained on 3 of them, and then extracted pre-logit layer from the classifier and fed into an LSTM. Classifier trained on 5 epochs each fold, 480 images with below pre-processing. Each epoch, each fold, we extract embedding layer (use TTA and avg embeddings) train a separate LSTM for 12 epochs on each of those - so 15 LSTMs (3 fold image models X 5 epochs), and average the predictions. Was a bit concerned the preprocessing filter may lose information, so trained the above again without the preprocessing filter and it did worse; but averaging both pipelines did ever so slightly better. The pipeline from first paragraph above would, for all intensive purposes be just as good as final solution, but as we needed to fix docu pre-stage 2 the two pipelines are in github and final solution.
Preprocessing:
- Used Appian’s windowing from dicom images. Linky
- Cut any black space. There were then headrest or machine artifacts in the image making the head much smaller than it could be - see visual above. These were generally thin lines, so used scipy.ndimage minimum_filter to try to wipe those thin lines. Linky
- Albumentations as mentioned in visual above.
Image classifier
- Resnext101 - did not spend a whole lot of time here as it ran so long. But tested SeResenext and Efficitentnetv0 and they did not work as well.
- Extract GAP layer at inference time Linky
Create Sequences
- Extract metadata from dicoms : Linky
- Sequence images on Patient, Study and Series - most sequences were between 24 and 60 images in length. Linky
LSTM
- Feed in the embeddings in sequence on above key - Patient, Study and Series - also concat on the deltas between current and previous/next embeddings ( and ) to give the model knowledge of changes around the image. Linky
- LSTM architecture lifted from the winners of first stage toxic competition. This is a beast - only improvements came from making the hiddens layers larger. Oh, we added on the embeddings to the lstm output and this helped a bit also. Linky
- For sequences of different length, padded them to same length, made a dummy embedding of zeros, and then through the results of this away before calculating loss and saving the predictions.
What did not help...
Too long to do justice... mixup on image, mixup on embedding, augmentations on sequences (partial sequences, reversed sequences), 1d convolutions for sequences (although SeuTao got it working)
Given more time
Make the classifier and the lstm model single end-to-end model.
Train all on stage2 data, we only got to train two folds of the image model on stage-2 data.
Note: Run environment with Docker file docker/RSNADOCKER.docker.
Note: The scripts below were run on an LSF cluster. This can be run outside of LSF, however within the above docker env, by just running the shell command within the double quotes. For eaxample, instead of bsub -app gpu -n =0 -env LSB_CONTAINER_IMAGE=darraghdog/kaggle:apex_build "cd /mydir && python3 myscript.py", just run, cd /mydir && python3 myscript.py.
-
A. Run script
download.shto download the data to/datafolder. For pretrained image weights we usetorchvision.models.resnetand the checkpointresnext101_32x8d_wsl_checkpoint.pth, taken from here. We also attach for your convenince.
B. This should also run a scripteda/window_meta2csv.pyto create meta data files. -
Create folds by executing
python eda/folds_v2.py -
Convert dicoms to jpeg by executing
eda/window_v1.py. Before doing this point the variables,path_imgwhere the dicom images are stored,path_dataat the meta file directories andpath_procto the directory to store the images which will be loaded to thetrainorigscript. -
A. Run training of resenext101 for 3 folds for 5 epochs by executing
sh scripts/resnext101v12/run_1final_train480.sh.
B. Run again training of resenext101 for 3 folds for 5 epochs by executingsh scripts/resnext101v13/run_1final_train480.sh. -
A. Extract embeddings for each of these runs (3 folds, 5 epochs) using
sh scripts/resnext101v12/run_2final_emb.sh. Note, this script uses test time augmentation, and extracts embeddings for the original image, horizontal flip and transpose.B. Extract embeddings for each of these runs (3 folds, 5 epochs) using
sh scripts/resnext101v13/run_2final_emb.sh. Note, this script uses test time augmentation, and extracts embeddings for the original image, horizontal flip and transpose. -
Train LSTM on image embeddings by sequencing the images per patient, series and study :
sh scripts/resnext101v13/run_3final_lstmdeltasum.sh -
Train LSTM on image embeddings by sequencing the images per patient, series and study :
sh scripts/resnext101v12/run_3final_lstmdeltasum.sh. -
Bag the results of each of the LSTM runs and create submission using
eda/val_lstm_short.py. Again, 3 folds 5 epochs, and then LSTM is bagged for the 12 epochs it runs. Here we simply average the results of all the last LSTM file outputs. submit2 -
For blending results.
9.1. Run another LSTM using
LSTM_submit_05.pyvia:sh izuit/LSTM_submit_05_f0.sh;sh izuit/LSTM_submit_05_f0.shandsh izuit/LSTM_submit_05_f0.sh.9.2. Bag results using
LSTM_submit_05_bagging.py9.3. Build final blend using
blend.ipynbwith alpha = .95 (best results for stage1 LB) submit1
Model (.scripts/ folder) |
Image Size | LSTM Epochs | Bag | TTA | Fold | Val | Stg1 Test | LB Public / Private | Comment |
|---|---|---|---|---|---|---|---|---|---|
| ResNeXt-101 32x8d retrained v12 fold 0 1 w/stg2 (v12&v13) with LSTM | 480 | 5 | LSTM 12X | v12 0 1 2- hflip transpose; v13 0 - hflip | 0 1 2 0 1 2 (v12 v13) | 0.05622, 0.05775, 0.05604, 0.05648, 0.05775, 0.05534 | --- | 0.697 / 0.044 | Incl stage 1 test, resnextv12/run_train1024lstmdeltattasum.sh resnextv12/run_train1024lstmdeltattasum.sh & eda/val_lstm_v22.py |
| ResNeXt-101 32x8d (v12&v13) with LSTM | 480 | 5 | LSTM 12X | v12 0 1 2- hflip transpose; v13 0 - hflip | 0 1 2 0 1 2 (v12 v13) | 0.05654, 0.05807, 0.05604, 0.05648, 0.05775, 0.05534 | 0.4544 | 0.654 / 0.045 | Incl stage 1 test, resnextv12/run_train1024lstmdeltattasum.sh resnextv12/run_train1024lstmdeltattasum.sh & eda/val_lstm_v22.py |
| ResNeXt-101 32x8d (v12&v13) with LSTM | 480 | 5 | LSTM 12X | v12 0 1 2- hflip transpose; v13 0 - hflip | 0 1 2 0 1 2 (v12 v13) | 0.05699, 0.05866, 0.05642, 0.05696, 0.05844, 0.05588 | 0.5703 | 0.675 / 0.045 | Excl stage 1 test, resnextv12/run_train1024lstmdeltattasum.sh resnextv12/run_train1024lstmdeltattasum.sh & eda/val_lstm_v21.py |
| ResNeXt-101 32x8d (v12&v13) with LSTM | 480 | 5 | LSTM 12X | v12 0 1 2- hflip transpose | 0 1 2 (v12) | 0.05699, 0.05866, 0.05642 | 0.5706 | 0.713 / 0.046 | Excl stage 1 test, resnextv12/run_train1024lstmdeltattasum.sh resnextv12/run_train1024lstmdeltattasum.sh & eda/val_lstm_v20.py |
Model (.scripts/ folder) |
Image Size | Epochs | Bag | TTA | Fold | Val | LB | Comment |
|---|---|---|---|---|---|---|---|---|
| ResNeXt-101 32x8d (v12&v13) with LSTM | 480 | 5, 5, 5, 5 | LSTM 12X | v12 0 1 2- hflip transpose; v13 0 - hflip | 0 1 2 0 (v13) | 0.05705 0.05866 0.05645 0.05690 | 0.057 | Hidden 2048, bag12 epochs, resnextv12/run_train1024lstmdeltattasum.sh resnextv12/run_train1024lstmdeltattasum.sh & eda/val_lstm_v14.py |
| ResNeXt-101 32x8d (v12&v13) with LSTM | 480 | 5, 5, 5, 5, 5 | LSTM 12X | v12 0 1 2- hflip transpose; v13 0 1 - hflip | 0 1 2 0 (v13) | 0.05687 0.05859 0.05651 0.05685 0.05839 | 0.057 | Hidden 2048, bag12 epochs, resnextv12/run_train1024lstmdeltattasum.sh resnextv12/run_train1024lstmdeltattasum.sh & eda/val_lstm_v16.py |
| ResNeXt-101 32x8d (v12) with LSTM | 480 | 5, 5, 5 | LSTM 12X | all folds 0-hflip, 1 2 - transpose | 0 1 2 | 0.05705 0.05866 0.05645 | 0.057 | Increase hidden units to 2048, bag12 epochs, scripts/resnextv12/run_train1024lstmdeltatta.sh & eda/val_lstm_v13.py , bsize 4 patients |
| ResNeXt-101 32x8d (v12) with LSTM | 480 | 5 | 9X | HFlip TTA on fold0 only | 0 1 2 | 0.05730 0.05899 0.05681 | 0.057 | Concat delta to prev and delta to next, bag9 epochs, scripts/resnextv12/trainlstmdelta.py & eda/val_lstm_v11.py , bsize 4 patients |
| ResNeXt-101 32x8d (v12) with LSTM | 480 | 6 | 5X | None | 0 | 0.0574 | 0.059 | Concat delta to prev and delta to next, bag4 epochs, scripts/resnextv12/trainlstmdelta.py, bsize 4 patients |
| ResNeXt-101 32x8d (v11) with LSTM | 384 | 5, 5, 6 | 5X | None | 0, 1, 2 | 0.05780, 0.05914, 0.05666 | 0.059 | 2X LSTM 1024 hidden units, bag4 epochs, scripts/resnextv11/trainlstmdeep.py & eda/val_lstm_v9.py, bsize 4 patients |
| ResNeXt-101 32x8d (v12) with LSTM | 480 | 5 | 5X | None | 0 | 0.05758 | 0.059 | 2X LSTM 1024 hidden units, bag4 epochs, scripts/resnextv12/trainlstmdeep.py, bsize 4 patients |
| ResNeXt-101 32x8d (v6) with LSTM | 384 | 7, 5, 7 | 6X, 4X, 6X | None | 0, 1, 2 | 0.5836, 0.6060, 0.5728 | 0.060 | 2X LSTM 256 hidden units, bag4 epochs, scripts/resnextv11/trainlstmdeep.py, bsize 4 patients |
| ResNeXt-101 32x8d (v11) with LSTM | 384 | 5 | 5X | None | 0 | 0.05780 | 0.060 | 2X LSTM 1024 hidden units, bag4 epochs, scripts/resnextv11/trainlstmdeep.py, bsize 4 patients |
| ResNeXt-101 32x8d (v6) with LSTM | 384 | 7 | 5X | None | 0 | 0.05811 | 0.061 | 2X LSTM 256 hidden units, bag4 epochs, scripts/resnextv6/trainlstmdeep.py, bsize 4 patients |
| SEResNeXt-50 32x8d (v3) with LSTM(1024HU) | 448 | 4 | 3X | None | 0, 1, 2, 3 | 0.05876, 0.06073, 0.05847, 0.06079 | 0.061 | 2X LSTM 1024 hidden units, bag8 epochs, scripts/resnextv6/trainlstmdeep.py, bsize 4 patients |
| ResNeXt-101 32x8d (v6) with LSTM | 384 | 7 | 3X | None | 0 | 0.05844 | 0.061 | 2X LSTM 256 hidden units, bag4 epochs, scripts/resnextv6/trainlstmdeep.py, bsize 4 patients |
| ResNeXt-101 32x8d (v8) with LSTM | 384 | 7 | 6X | None | 0 | ----- | 0.062 | 2X LSTM 256 hidden units, bag4 epochs, scripts/resnextv8/trainlstmdeep.py, bsize 4 patients |
| ResNeXt-101 32x8d (v4) with LSTM | 256 | 7 | 5X | None | 0 | 0.06119 | 0.064 | 2X LSTM 256 hidden units, bag4 epochs, scripts/resnextv4/trainlstmdeep.py, bsize 4 patients |
| ResNeXt-101 32x8d (v4) with LSTM | 256 | 7 | 5X | None | 0 | 0.06217 | 0.065 | LSTM 64 hidden units, bag 5 epochs, scripts/resnextv4/trainlstm.py, bsize 4 patients |
| ResNeXt-101 32x8d (v8) | 384 | 7 | 5X | None | 5 (all) | ----- | 0.066 | Weighted [0.6, 1.8, 0.6] rolling mean win3, transpose, submission_v6.py, bsize 128 |
| ResNeXt-101 32x8d (v8) | 384 | 7 | 4X | None | 5 (all) | ----- | 0.067 | Weighted [0.6, 1.8, 0.6] rolling mean win3, transpose, submission_v6.py, bsize 128 |
| ResNeXt-101 32x8d (v6) | 384 | 7 | 5X | None | 0 | 0.06336 | 0.068 | Weighted [0.6, 1.8, 0.6] rolling mean win3, transpose, submission_v5.py, bsize 32 |
| ResNeXt-101 32x8d (v4) | 256 | 7 | 5X | None | 0 | 0.06489 | 0.070 | Weighted [0.6, 1.8, 0.6] rolling mean win3, transpose, submission_v4.py, bsize 64 |
| ResNeXt-101 32x8d (v4) | 256 | 7 | 5X | None | 0 | 0.06582 | 0.070 | Rolling mean window 3, transpose, submission_v3.py, bsize 64 |
| ResNeXt-101 32x8d (v4) | 256 | 4 | 3X | None | 0 | 0.06874 | 0.074 | Rolling mean window 3, transpose, submission_v3.py, bsize 64 |
| EfficientnetV0 (v8) | 256 | 6 | 3X | None | 0 | 0.07416 | 0.081 | Rolling mean window 3, no transpose, submission_v2.py, bsize 64 |
| EfficientnetV0 (v8) | 384 | 4 | 2X | None | 0 | 0.07661 | 0.085 | With transpose augmentation |
| LSTM on logits from ResNeXt-101 32x8d (v4) | 256 | 3 | 3X | None | 0 | 0.063 | 0.082 | LSTM on sequence of patients logits, bsize 4 patients |
| EfficientnetV0 (v8) | 384 | 2 | 1X | None | 0 | 0.07931 | 0.088 | With transpose augmentation |
| EfficientnetV0 (v8) | 384 | 11 | 2X | None | 0 | 0.08330 | 0.093 | With transpose augmentation |
| EfficientnetV0 | 224 | 4 | 2X | None | 0 | 0.08047 | ???? | Without transpose augmentation |
| EfficientnetV0 | 224 | 4 | 2X | None | 0 | 0.08267 | ???? | With transpose augmentation |
| EfficientnetV0 | 224 | 2 | 1X | None | 0 | 0.08519 | ???? | With transpose augmentation |
| EfficientnetV0 | 224 | 11 | 2X | None | 0 | 0.08607 | ???? | With transpose augmentation |