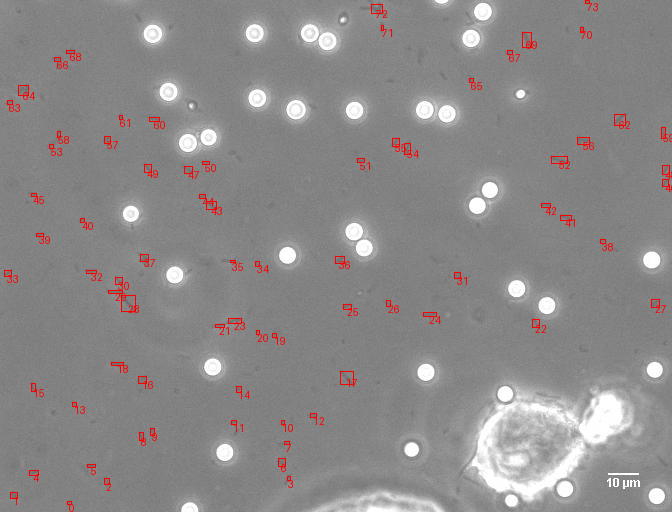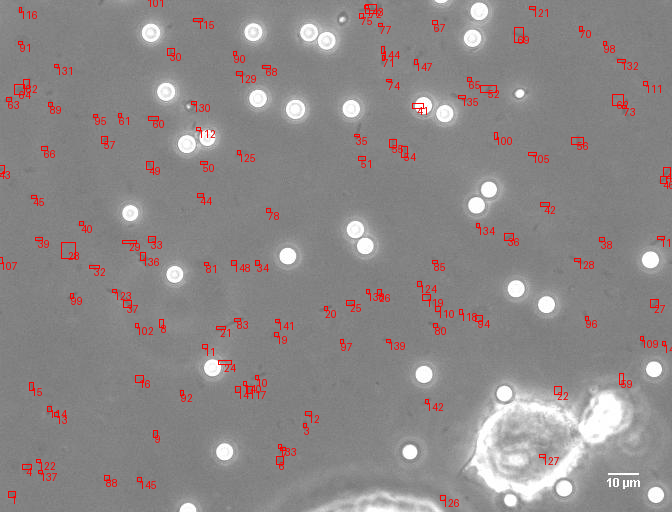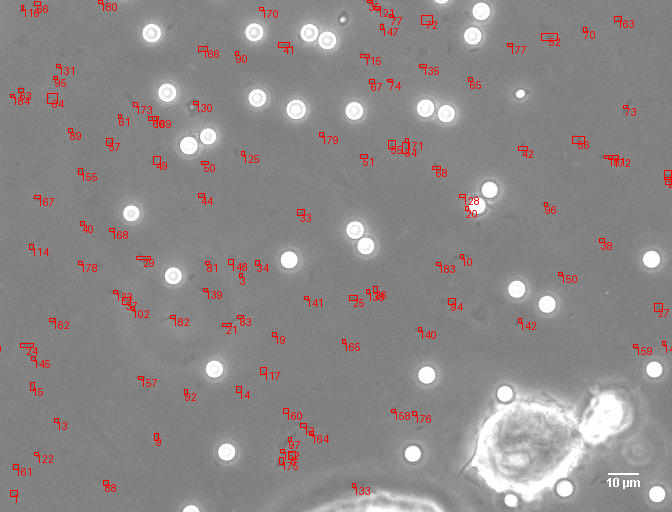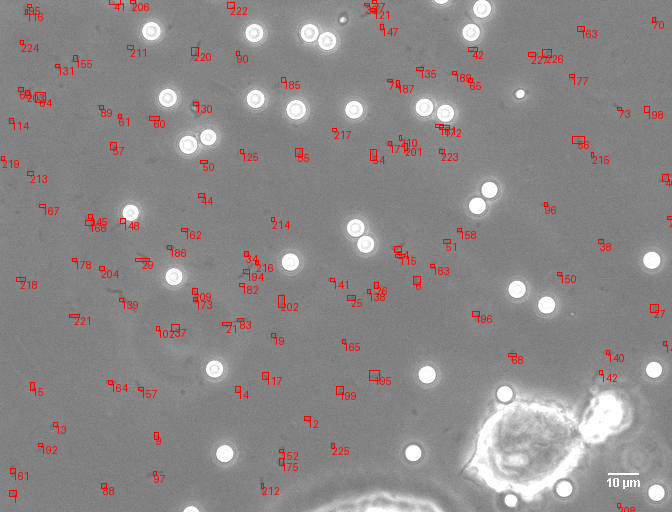
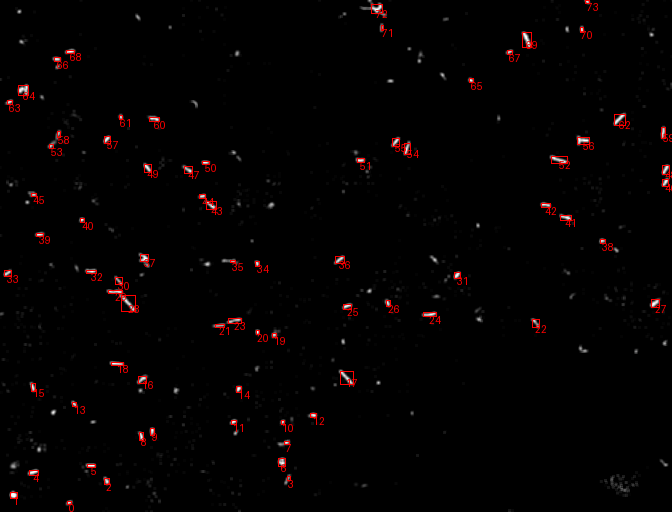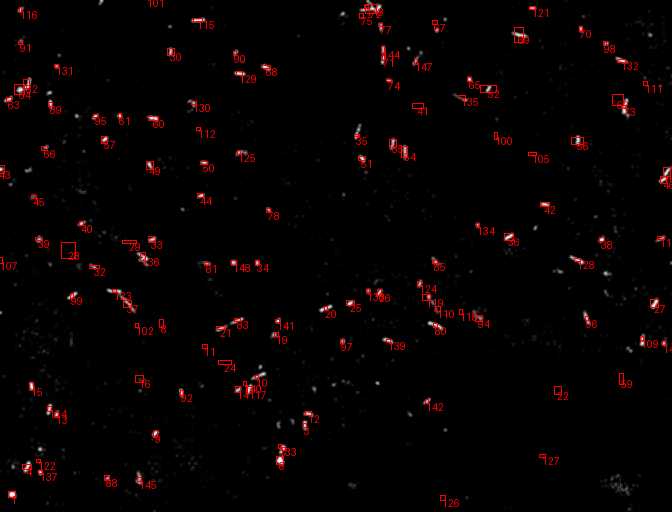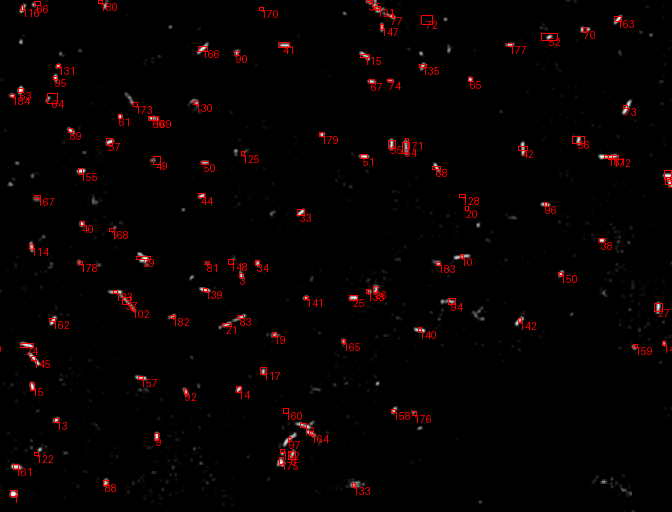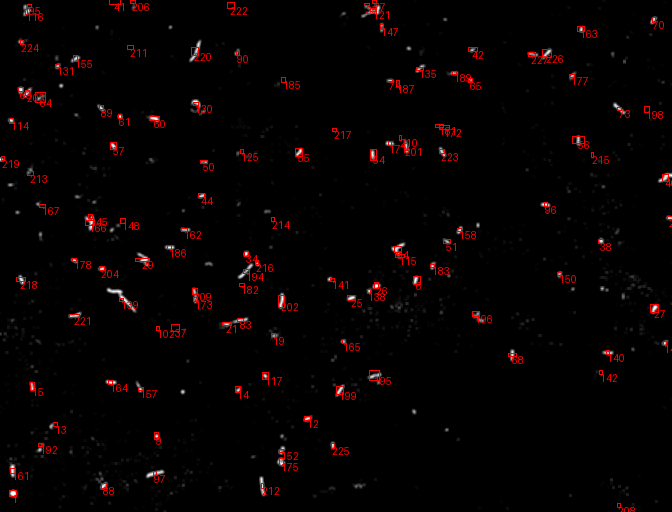
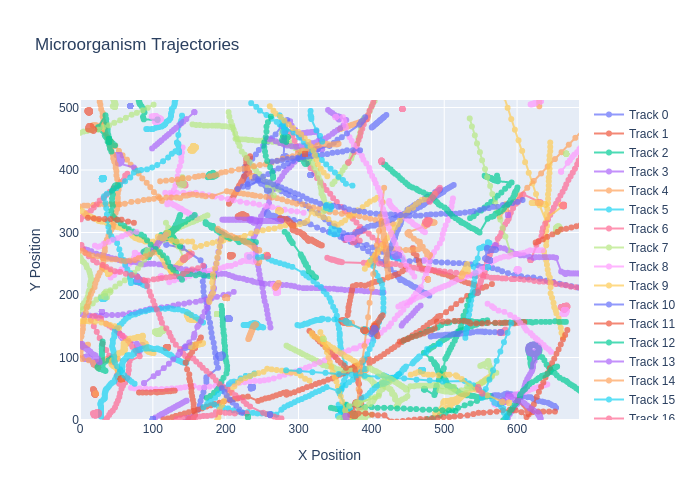
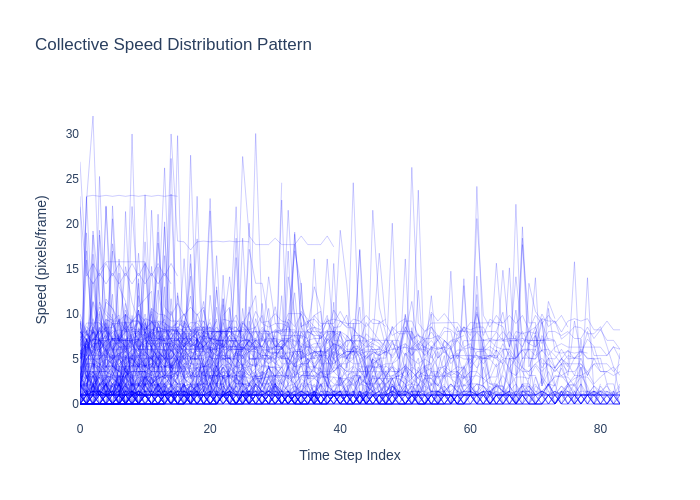
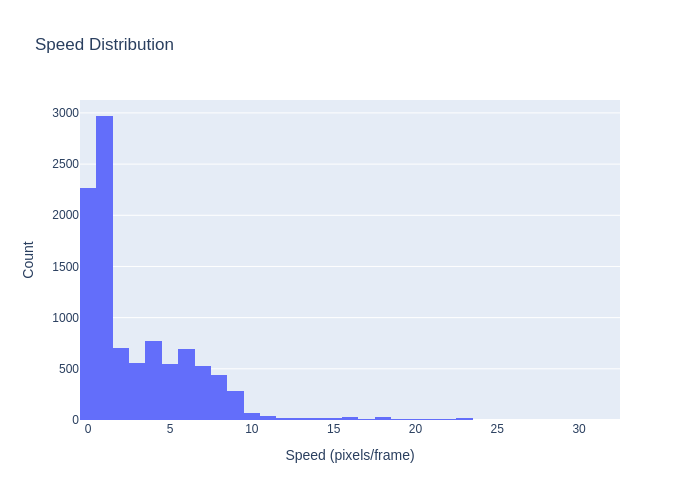
A comprehensive computer vision pipeline for analyzing microorganism 🦠 motility patterns and diffusion characteristics.
Deadline: 28th February 2025
- Multi-modal Tracking
- Optical flow-based motion estimation
- Kalman-filter enhanced object detection
- Synthetic trajectory generation
- Advanced Diffusion Analysis
- 6 statistical hypothesis tests for motility patterns
- Automated trajectory classification
- Comprehensive visualization toolkit
- Scalable Architecture
- Modular pipeline design
- Configurable hypothesis parameters
- Batch processing capabilities
Our framework implements sophisticated statistical tests to characterize microbial motion:
| Hypothesis Test | Key Mechanism | Biological Relevance |
|---|---|---|
Circular Motion CircularMotionTest
|
Curvature analysis + rotation angle detection | Magnetotactic bacteria, spiral-seeking organisms |
Directional Switching DirectionalSwitchTest
|
Fourier analysis of angular changes | Run-and-tumble motility (e.g., E. coli) |
Gaussian Random Walk GaussianRandomWalkTest
|
Kolmogorov-Smirnov normality test | Passive diffusion in isotropic environments |
MSD Linear Fit MSDLinearTest
|
R² evaluation of mean squared displacement | Normal diffusion processes |
Persistent Motion PersistentMotionTest
|
Velocity autocorrelation analysis | Active transport mechanisms |
Subdiffusion SubDiffusionTest
|
Power law exponent ( |
Crowded environments, viscoelastic media |
# Clone repository
git clone git@github.com:Ophiase/Microorganism-Computer-Vision.git
cd Microorganism-Computer-Vision
# Install dependencies
make pip
# Download sample dataset
make extractProcess real videos:
# Full pipeline execution
make transform # Preprocess videos
make detection # Track microorganisms
make render # Generate gifs
make analysis # Run statistical tests and graphicsGenerate synthetic data:
make synthetic # Create trajectory datasets
python3 -m script.main --task analysis --video synthetic_brownian
# "brownian" can be replaced by directed, sinusoidal, confined, ctrw, ...Key Makefile Targets:
detection # Track objects in videos
render # Generate trajectory visualizations
analysis # Perform statistical testing
synthetic # Generate synthetic trajectoriesmicroorganism-cv/
├── data/ # Raw and processed data
│ ├── preprocessed/ # Normalized video tensors
│ └── tracking/ # Trajectory datasets
├── logic/ # Core algorithms
│ ├── diffusion/ # Statistical tests
│ ├── filters/ # Image processing
│ └── structure/ # Data models
├── script/ # Pipeline components
└── visualization/ # Plotting utilitiesOur pipeline implements a multi-stage analysis process:
- Optical flow estimation
- Spatial normalization
- Noise reduction
- Adaptive thresholding (❌ not implemented)
- Connected component analysis
- Kalman-filter based tracking
- MSD calculations
- Velocity autocorrelation
- Directional persistence metrics
- Hypothesis testing
- Confidence interval estimation (❌ not implemented)
- Motility pattern classification