Prostate mpMRI lesion detection system
This repository contains the code for the paper Deep Learning system for fully automatic detection, segmentation and Gleason Grade estimation of prostate cancer in multiparametric Magnetic Resonance Images (currently in preprint), which proposes a fully automatic system that takes prostate multi-parametric magnetic resonance images (mpMRIs) from a prostate cancer (PCa) suspect and, by leveraging the Retina U-Net detection model, locates PCa lesions, segments them, and predicts their most likely Gleason grade group (GGG).
This model has been adapated to only use ProstateX data, achieving an AUC of 0.87 at the challenge online learderboard, hence tying up with the winner of the original ProstateX challenge.
Please, cite the peer-reviewed paper if you use any of the code in this repository:
Oscar J. Pellicer-Valero, José L. Marenco Jiménez, Victor Gonzalez-Perez, Juan Luis Casanova Ramón-Borja, Isabel Martín García, María Barrios Benito, Paula Pelechano Gómez, José Rubio-Briones, María José Rupérez, José D. Martín-Guerrero, Deep Learning for fully automatic detection, segmentation, and Gleason Grade estimation of prostate cancer in multiparametric Magnetic Resonance Images. Scientific Reports. February, 2022.
This is an example of the output of the model:
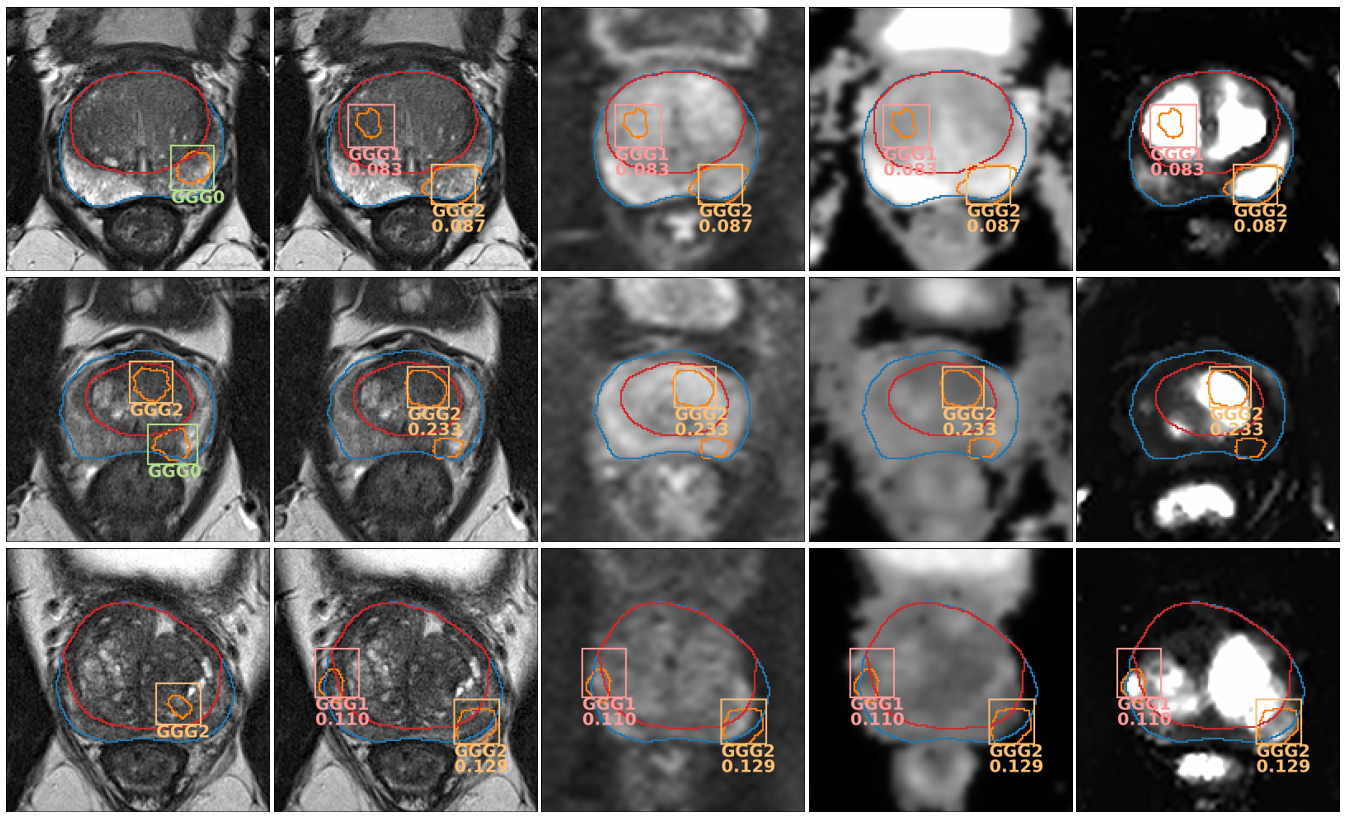
Output of the model evaluated on three ProstateX test patients. First image from the left shows the GT on the T2 mpMRI sequence; the rest show the output predictions of the model on different sequences (from left to right: T2, b800, ADC, Ktrans). GGG0 (benign) detections are not shown and only the highest-scoring detection is shown for highly overlapped detections (IoU > 0.25). Detections with a confidence below the lesion-wise maximum sensitivity setting (t=0.028) are also ignored.
Overview
This repository contains three main Jupyter Notebooks:
- ProstateX preprocessing: Performs the preprocssing of the ProstateX data. All steps, from downloading the data to configuring the Notebook are explained within it. At the end, it creates a
ID_img.npyfile containng the processed image and masks, aID_rois.npyfile containing the lesions, and ameta_info_ID.picklecontaining important meta information for every patientID. - Registration example: Notebook for performing general medical image registration, but adapated to register prostate ADC maps to T2 sequences. It takes some unregistered
ID_img.npyfiles and produces a SimpleITK transform for each of them as output asID.tfm. - Result analysis: It analyzes the results produced by the Retina U-Net model (
processed_pred_boxes_overall_hold_out_list_test.pickle), allowing to plot the ground truth alongside the predicted detections, obtain metrics and ROC curves at lesion and patient level, and generate a ProstateX challenge submission.
Note: the Notebooks should not be run within Jupyter Lab, but rather opened intedependently in Jupyter Notebook, otherwise the plot_lib visualizations might not display properly. For more information on the issue, see this link.
Additionally, the directory ./MDT_ProstateX contains a complete fork of the Medical Detection Toolkit, which is employed as the backend for the system and has been modified to adapt it to this task. All modifications to any of the files have been listed in their headers, in compliance with the Apache 2.0 license used by that project.
Installation
To install, please clone this repository and install required packages. It is recommended to use a package manager such as pip or conda (conda was the only one tested, so use pip at your own risk). If not sure, you can download and install the latest miniconda release before continuing and install git from the conda console: conda install git
git clone https://github.com/OscarPellicer/prostate_lesion_detection.git
#You probably want to create an environment first. Using conda:
conda create -n prostate_lesion python=3.7
conda activate prostate_lesion
#Install required libraries. Using conda:
conda install matplotlib numpy ipywidgets ipython scipy pandas==0.25.3 jupyter ipython scikit-learn
conda install SimpleITK==1.2.4 -c simpleitk
conda install pydicom -c conda-forge
#pip install matplotlib numpy ipywidgets ipython scipy simpleitk==1.2.4 pandas==0.25.3 pydicom jupyter ipython scikit-learnYou will also need plot_lib for plotting the mpMRIs within the Jupyter Notebooks. To install it, you may simply clone the repository to your home path:
git clone https://github.com/OscarPellicer/plot_lib.gitNow we navigate to the cloned repository:
cd prostate_lesion_detection/Then, some zip files will need to be unpacked, in particular:
ProstateX_masks.zip: This contains automatically generated ProstateX masks for the whole prostate as well as the central zone, using the model from: Robust Resolution-Enhanced Prostate Segmentation in Magnetic Resonance and Ultrasound Images through Convolutional Neural NetworksProstateX_transforms.zip: This contains the transforms for registration of the ProstateX dataset. These were generated using the Notebook Registration example from this repository../MDT_ProstateX/experiments/exp0/test/test_boxes.zip: This is the output of the model for the test set, which can be analyzed using the Notebook Result analysis from this repository../MDT_ProstateX/experiments/exp0/test/train_boxes.zip: This is the output of the model for the train set, which can be used to compete in the ProstateX online challenge using the Notebook Result analysis from this repository.
All these can be manually unziped, or using the following commands
unzip \*.zip -d ./
unzip ./MDT_ProstateX/experiments/exp0/test/\*.zip -d ./MDT_ProstateX/experiments/exp0/test/At this point, you should be able to run all the provided Jupyter Notebooks, but you will not be able to use the Medical Detection Toolkit for training or inference yet, as you still need to install the packages required by it. In summary, you will need to do two things (make sure to run everything from within the prostate_lesion conda / pip environment).
Note: If you have any doubts in this step, it might be worth looking at the original documentation of the toolkit (README), alhtough some things here differ slightly. For instance, here we use pytorch 1.7, instead of torch 1.4
First, install pytorch:
conda install pytorch==1.7.0 -c pytorchThen, you need to go to https://developer.nvidia.com/cuda-gpus, look at the Compute Capability of your Nvidia GPU, and create an environmental variable with the latest version that your GPU supports. This is required for the correct compilation of some CUDA custom extensions. E.g.:
export TORCH_CUDA_ARCH_LIST="6.0"Please note that a CUDA compiler should be already installed in the system for the next step to work, which can be checked by running which nvcc, which should return the install path of nvcc, or nothing if nvcc is not installed.
Finally, go to the MDT_ProstateX folder, and run the setup.py script to install the rest of required libraries and build the custom extensions:
cd MDT_ProstateX
python setup.py installUsage
All the provided Jupyter Notebooks can be run without actually installing the Medical Detection Toolkit requirements.
To use the provided model for inference on new data, you will have to first preprocess your images identicaly to how the ProstateX images have been processed using ProstateX preprocessing. Then, replace the IDs in the test key of the ss_v2 dictionary at the beginning of the file ./MDT_ProstateX/experiments/exp0/data_loader.py by your own IDs. Finally, run the model in inference mode and aggragate the results:
python exec.py --mode test --exp_source experiments/exp0 --exp_dir experiments/exp0
python exec.py --mode analysis --exp_source experiments/exp0 --exp_dir experiments/exp0To use the Medical Detection Toolkit for training, please create a directory within the MDT_ProstateX/experiments directory containing a copy of the files: configs.py, custom_transform.py, and data_loader.py from the provided experiment MDT_ProstateX/experiments/exp0. These files will have to be modified to fit your needs, or, at the very least, the ss_v2 dictionary at the beginning of the file data_loader.py should be modified to include your own IDs. Then, to run the model and produce the predictions on your test set:
python exec.py --mode train_test --exp_source experiments/exp1 --exp_dir experiments/exp1
python exec.py --mode analysis --exp_source experiments/exp1 --exp_dir experiments/exp1Either if you use the model for inference or training on your data, you will be able to analyze the results by using the Result analysis Notebook. You must remember to change the value of the variable SUBSET in this Notebook to point to the new predictions.
Contact
If you have any problems, please check further instructions in each of the provided Notebooks, create a new Issue, or directly email me at Oscar.Pellicer at uv.es