Title: Deep Learning for Wrist Fracture Detection: Are We There Yet?
arXiv link
A transfer learning pipeline to detect wrist fracture from DICOM files. It has two blocks: Landmark Localization Block
and Fracture Detection Block.
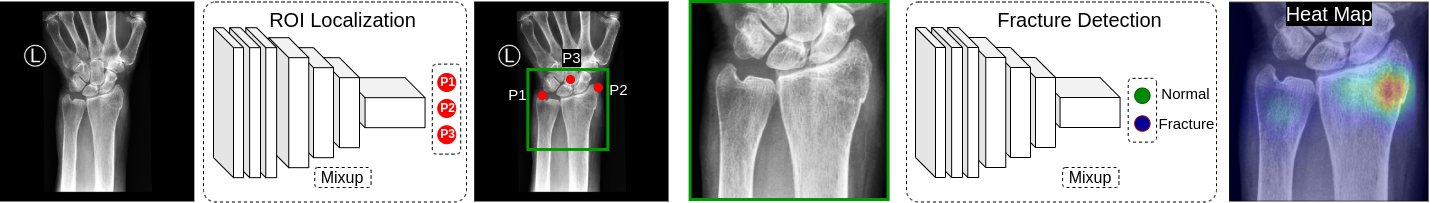
Both of the blocks are yml configuration based. We used OmegaConf for this purpose. Each executable python file can
either run standalone or requires a yml file as experiment argument to be passed down at command line.
Landmark Localization Block is adapted from KNEEL. However, we developed some data
augmentation methods suited to our task. The localizer folder contains the source code for Landmark Localizer and
structured as
localizer
|---config
| |---experiment
|---kneel_before_wrist
| |---data
| |---model
|---scripts
The config folder contains the initial default configuration and a configuration processor. There is a folder named
experiment inside config folder which holds confgiguration for different experiments.
kneel_before_wrist hosts the body of the localizer part of our pipeline. It has two sub-directory: data and model.
The data folder contains utilities necessary to process and augment data for training and evaluation. model
sub-directory contains the pytorch lightning version of HourGlass network which we will use for training the localizer.
The scripts directory hosts all the experiment scripts for which the yaml configurations are created.
-
First step for training your own localizer is to collect data. For our research we used private hospital data, threfore it cannot be shared. To make things simple, we used csv file to store meta data about the dataset. This way, you don't have to load the full dataset to the memory rather fecth the file location from csv file and read it just-in-time. So, we are dealing with wrist fracture images. We will consider posterioanterio (PA) and lateral (LAT) view of the wrist x-ray. To make your own dataset, you have to create a csv metadata file containing at least
Fname, Points, Sidecolumns.Fnameis the absolute path to the wrist image,Pointscolumn will contain the landmark coordinates of top of distal ulna, top of distal radius and assumed center of the wrist for PA view and two distinguishalbe points on top part of distal radio-ulna bone and the assumed center of wrist for LAT view. As the name suggest, theSidecolumn contains the side information of corresponding wrist x-ray. Put 0 for PA and 1 for LAT. Once the metadata is ready, we can move forward. -
Second step is to clone
wrist_landmark.yamlconfiguration file and modify the clone. Inside the yaml file modify following
data_home: # root folder that contains the data folder
data_folder: # your data folder name
meta: the csv meta file you have created. should be inside data folder
- Once you are done with step 2, run the
train_ptl.py --experiment=YourClonedYAMLFile. This file is located insidescriptsfolder. It will start the training.
The classifier folder hosts the Fracture Detection Block. It has a similar structure like localizer.
classifier
|---config
|---fracture_detector
| |---callback
| |---data
| |---model
|---script
Like before config folder hosts the script configurations. fracture_detector folder hosts necessary folders and
files for model, data and training related stuffs. Inside this folder, there are three folders: 1) callback (hosts
callback function definitions), 2) data (hosts data related utilities) and 3) model (hosts model definition and
training methods)
Step 1. First step to train your custom fracture detector is to collect data using the localizer model trained previously.
Save the generated ROI with the corresponding ID as filename. Create a csv metadata file with ID, Side, Fname(optional)
and Fracture columns. Say, the meta file name is your_meta.csvCreate a root folder which we will use as data home where the generated ROI images and the csv
meta file are saved. There shoudl be PA and LAT folder in the root folder to host PA ROI and LAT ROI respectively.
Step 2. Clone the existing training conf fracture_detector_seresnet.yaml to your_config_file.yaml.
Open your_config_file.yaml and update the following field
data_home: root
meta: your_meta.csv
Step 3. Once you are done with the config file go inside the scripts folder and run python train_ptl.py experiment=your_config_file.
this will start the training.
Step 1. Create a csv meta file of for the data you want to predict. Use ID, Side, Fname, and Fracture columns.
Step2. Clone fracture_deteciton_testset_1.yaml to your_testset.yaml
Step 3. Find and update the following field in your_testset.yaml
dataset:
train_data_home: root
test_data_home: /location/of/test/data
meta: /absolute/location/to/your_testset.csv
save_path: /absolute/location/to/save/prediction.csv
snapshot_folder: /folder/location/where/fracture/detector/models/are/saved
save_image: true or false
save_image_dir: /folder/location/if/you/want/to/save/output/images
localizer:
snapshot_folder: /folder/location/where/roi/localizer/models/are/saved
dataset:
train_data_home: /folder/location/where/localization/data/are/stored
keep only Fracture in gt. If you want to save gradcam set save_gradcam: true and define gradcam_dir
Step 4. Now in the scripts folder run, python test.py experiment=your_testset
This will do inference on your data, the predicitons will be saved in the csv file you defined.
Use the following commands to get the trained models.
wget http://mipt-ml.oulu.fi/models/DeepWrist/Fracture_Detection_Block.tar.gz
wget http://mipt-ml.oulu.fi/models/DeepWrist/ROI_Localization_Block.tar.gz
For citation, please use the following bibtex
@misc{raisuddin2020deep,
title={Deep Learning for Wrist Fracture Detection: Are We There Yet?},
author={Abu Mohammed Raisuddin and Elias Vaattovaara and Mika Nevalainen and Marko Nikki and Elina Järvenpää and Kaisa Makkonen and Pekka Pinola and Tuula Palsio and Arttu Niemensivu and Osmo Tervonen and Aleksei Tiulpin},
year={2020},
eprint={2012.02577},
archivePrefix={arXiv},
primaryClass={cs.CV}
}