1- Run this command:
# Choose the pipeline version to install
export PIPELINE_VERSION=1.8.4
# Run the setup cluster script
chmod +x deployment/local/setup-cluster.sh
./setup-cluster.sh- This will download
kubectl,k3sand install kubeflowCRDSandKubeflow pipelines. - If you want to install a different version of kubeflow pipelines just update the environment variable
PIPELINE_VERSION
2- Verify setup by running:
kubectl get pods -n kubeflow3- Open Kubeflow UI:
kubectl port-forward svc/ml-pipeline-ui 8080:80 -n kubeflow- Pipeline ui is available at http://localhost:8080
-
Setup directly on the host (optionally using
venv):virtualenv venv source venv/bin/activate pip install -r requirements.txt pip install -e .
-
(Optional) Build Docker images and push them to your preffered registry (
e.g: REGISTRY=your_regitry)# Build Image for data ingestion module docker build -t $REGISTRY/biobb_io:3.6.0 -f biobb_md_setup/components/data_ingestion/Dockerfile . # Build Image for chemical conversion module docker build -t $REGISTRY/biobb_chemistry:3.6.0 -f biobb_md_setup/components/chemical_conversion/Dockerfile . # Build Image for molecular dynamics analysis module docker build -t $REGISTRY/biobb_analysis:3.7.0 -f biobb_md_setup/components/md_analysis/Dockerfile . # Build Image for md simulation using amber module docker build -t $REGISTRY/biobb_amber:3.6.2 -f biobb_md_setup/components/md_simulation/Dockerfile . # Build Image for all related pdb operations (extraction, modification) module docker build -t $REGISTRY/biobb_structure_utils:3.6.1 -f biobb_md_setup/components/pdb_operations/Dockerfile .
-
If rebuilt, this images needs to be pushed and then updated in the
biobb_md_setup/pipeline/pipeline.pyfile. -
For Demo purposes This images has been already built and pushed to
DockerHub:
docker pull oussemalouati/biobb_structure_utils:3.6 docker pull oussemalouati/biobb_amber:3.6.2 docker pull oussemalouati/biobb_chemistry:3.6.0 docker pull oussemalouati/biobb_io:3.6.0 docker pull oussemalouati/biobb_analysis:3.7.0
-
-
Compile the pipeline and generate the YAML template.
python biobb_md_setup/pipeline/pipeline.py
- This will generate
biobb_md_setup/pipeline/pipeline.yaml - Use this
biobb_md_setup/pipeline/pipeline.yamlto create new pipeline using the Kubeflow UI
- This will generate
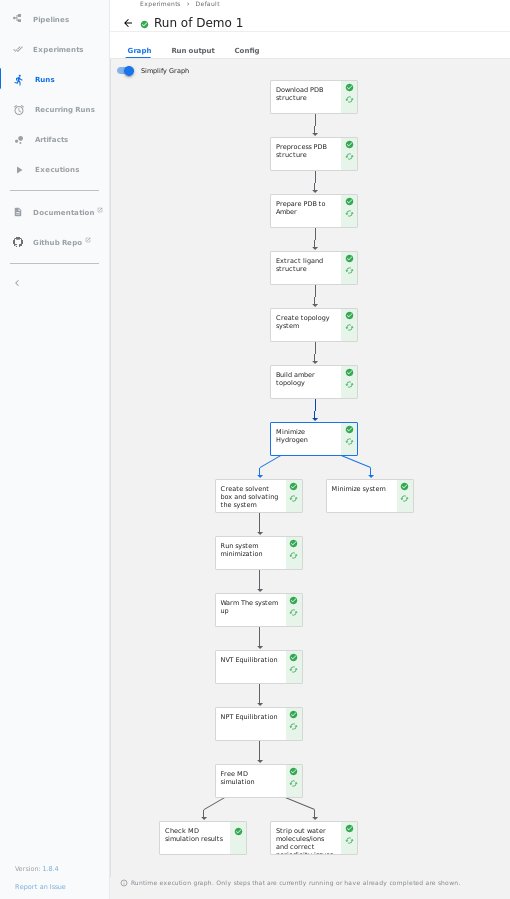
- Full Pipeline:
- All plotly figures from the notebook can been seen in the
visualisationtab in Kubeflow UI in their corresponding steps
The codebase is structured as follows:
.
|-- deployment ................. # Folder hosting all deployment related scripts
| `-- local ................... # Scripts for local deployment environment
|-- notebooks ................. # Notebooks
`-- biobb_md_setup .............. # Main folder
|-- components ................ # System component implementations
| |-- chemical_conversion ..... # Module for chemical conversion related scripts
| |-- data_ingestion ............. # Data ingestion module
| |-- data_visualisation ......... # Data visualisation module
| |-- md_analysis ................ # Molecular dynamics analysis module
| |-- md_simulation .............. # Module to gather all molecular dynamics using amber tools
| `-- pdb_operations ............. # Module for pdb operations (extractions, modifications)
|-- pipeline ................ # Pipeline in Kubeflow
| `-- pipeline.py
`-- version.py ................ # package version
`-- .gitignore .................... # Git untracked files
|
├── .pre-commit-config.yaml ....... # Git pre-commit webhooks for linting and testing
│
├── setup.py ...................... # package setup script
│
├── CONTRIBUTING.md .............. # Project contribution guidelines
├── LICENSE ....................... # Project software license
└── README.md ..................... # This document-
Run:
virtualenv venv source venv/bin/activate pip install -r requirements.txt pip install . python biobb_md_setup/pipeline/pipeline.py
-
Upload the generated
biobb_md_setup/pipeline/pipeline.yamlto your Kubeflow UI -
Wait for results
-
The pipeline code base is divided into
5logical sections as shown underbiobb_md_setup/components -
5different dockerfiles are used that are built on top of their correspondingquay.io/biocontainers/*. For some of them we are not actually adding any external/internal dependencies, but it is a good practice first to not rely on public images, second to be able at any time to extend the image by adding new dependecies or code. -
Instead of the
5Dockerfiles we can have a single big Dockerfile with all dependecies inside.-
Advantages:
- Not having to deal with multiple dockerfiles
-
Disadvantages:
-
This docker images will have a size
~5GB, the other 5 images sizes are:oussemalouati/biobb_structure_utils .. 333MB
oussemalouati/biobb_amber ............ 1.43GB
oussemalouati/biobb_analysis ......... 1.51GB
oussemalouati/biobb_chemistry ........ 1.22GB
oussemalouati/biobb_io ............... 330MB\Pulling an image of 5GB each time just to download a PDB or do a chemical conversion is an added overhead and will create unessary latency in each pipeline
-
condainside Docker is known to have multiple issues -
It is a good practice to divide our code base to different components that are each dedicated to just certain logical task, For better debugging, easier dependecy management.
e.g:biobbpackages used supportpython 3.7as the latest version, so in the future if we ever have a new components that needspython 3.8or later, the system is already modular we can add new components with it's own dockerfile without creating conflict.
-
-
-
Components in this pipeline are created based on
3criterias- Consecutive steps that do the same logical function (
e.g: molecular dynamics related scripts) - Consecutive steps that uses the same base image (
e.g: Amber) - (If a plot exists) Steps finishes with a plotly plot if it exists
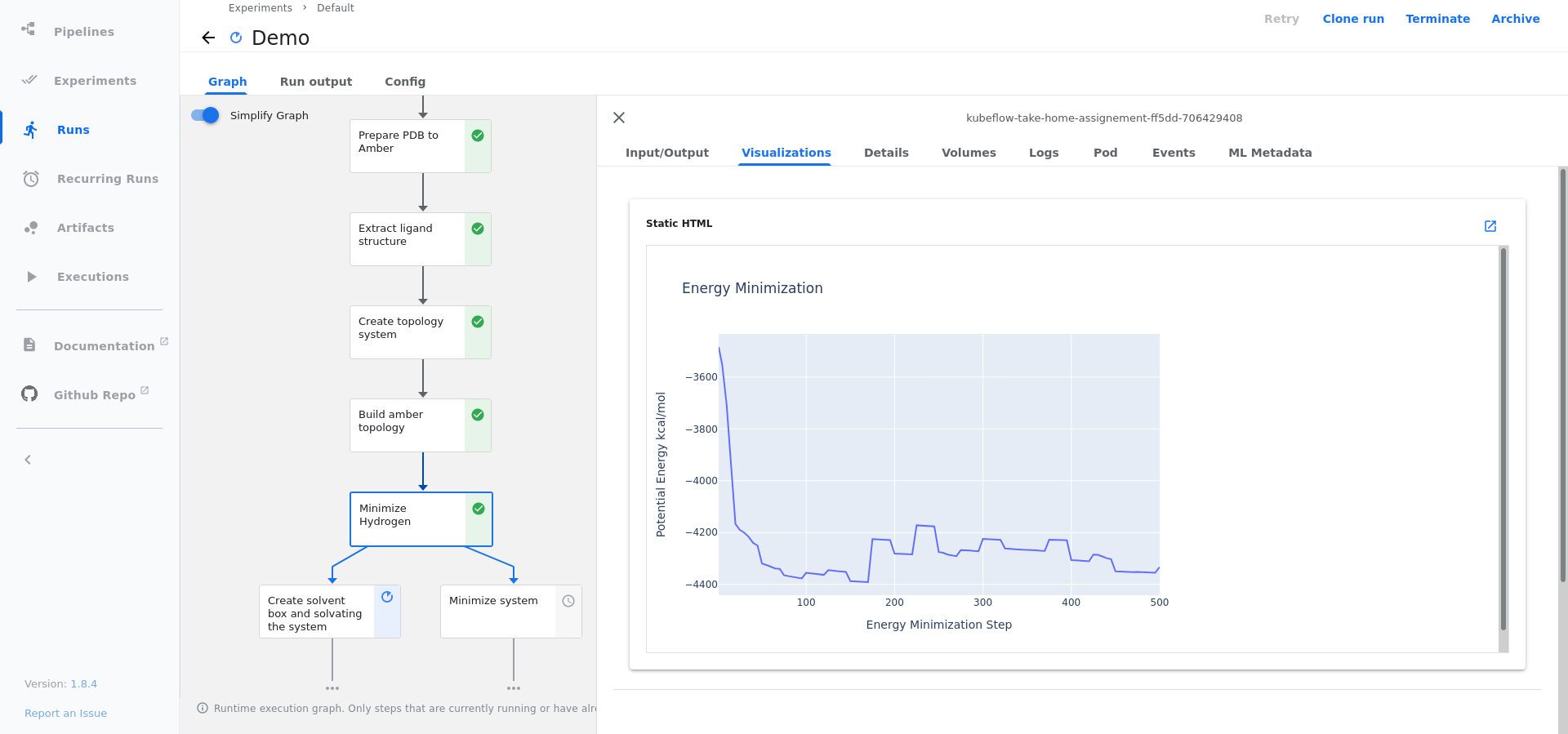
Example:
- Step 1: Hydrogen minimization
- Step 2: Checking Hydrogen minimization
- Step 3: Plot Energy minimization
--> These three steps are part of the molecular dynamics process (
Criteria 1) and uses the same base image (Criteria 2) and finishes with a plotly plot (Criteria 3) So these 3 steps are to be run together inside1Kubeflow step as shown in the screenshot above, the step is calledMinimize Hydrogen. - Consecutive steps that do the same logical function (
- Disadvantage: With kubeflow we aren't able to have interactive plots
-
Add an object storage (GSC, S3, MinIO) to store the final output files
-
Add Logging
-
Add unit tests
-
Improve code Documentation
-
Add the code to a code hosting platform like
Github, and create a CI pipeline to run steps likecode linting, run differenttests,Buildandpushthe different images