Flowdec is a library containing TensorFlow (TF) implementations of image and signal deconvolution algorithms. Currently, only Richardson-Lucy Deconvolution has been implemented but others may come in the future.
Flowdec is designed to construct and execute TF graphs in python as well as use frozen, exported graphs from other languages (e.g. Java).
Here are a few other features, advantages, and disadvantages of the project currently:
Highlights
- Support for Windows, Mac, and Linux - Because TensorFlow can run on these platforms, so can Flowdec.
- Client Support for Java, Go, C++, and Python - Using Flowdec graphs from Python and Java has been tested, but theoretically they could also be used by any TensorFlow API Client Libraries.
- Point Spread Functions - PSFs can be defined as json configuration files to be generated dynamically during the deconvolution process using a Fast Gibson-Lanni Approximation Model (which can also create Born & Wolf kernels as a degenerate case).
- GPU Accleration - Executing TensorFlow graphs on GPUs is trivial and will happen by default w/ Flowdec if you meet all of the TensorFlow requirements for this (i.e. CUDA Toolkit installed, Nvidia drivers, etc.).
- Performance - There are other open source and commercial deconvolution libraries that run with partial GPU acceleration, which generally means that only FFT and iFFT operations run on GPUs while all other operations run on the CPU. For example, on a roughly 1000x1000x11 3D volume with a PSF of the same dimensions this means that execution times look like:
- CPU-only solutions: 10 minutes
- Other solutions with FFT/iFFT GPU acceleration: ~40 seconds
- Flowdec/TensorFlow with full GPU acceleration: ~1 second
- Signal Dimensions - Flowdec can support 1, 2, or 3 dimensional images/signals.
- Multi-GPU Usage - This has yet to be tested, but theoretically this is possible since TF can do it (and this Multi-GPU Example is a start).
- Image Preprocessing - A trickier part of deconvolution implementations is dealing with image padding and cropping necessary to use faster FFT implementations -- in Flowdec, image padding using the reflection of the image along each axis can be specified manually or by letting it automatically round up and pad to the nearest power of 2 (which will enable use of faster Cooley-Tukey algorithm instead of the Bluestein algorithm provided by Nvidia cuFFT used by TF).
- Visualizing Iterations - Another difficulty with iterative deconvolution algorithms is in determining when they should stop. With Richardson Lucy, this is usually done somewhat subjectively based on visualizing results for different iteration counts and Flowdec at least helps with this by letting
observerfunctions be given that take intermediate results of the deconvolution process to be written out to image sequences or stacks for manual inspection. Future work may include using Tensorboard to do this instead but for now, it has been difficult to get image summaries working within TF "while" loops.
Disadvantages
- No GUIs - Flowdec is intended for use by those familiar with programming but some future work might include an ImageJ plugin (if there's interest in that). For those looking for something more interactive, imagej-ops is likely your best bet which currently supports the same PSF generation model used in Flowdec as well as Richardson Lucy deconvolution. At the moment it doesn't include full GPU acceleration but that may be on the way as part of imagej-ops-experiments. See this github issue for more details.
- No Blind Deconvolution - Currently, nothing in this arena has been attempted but since much recent research on this subject is centered around solutions in deep learning, TensorFlow will hopefully make for a good platform in the future.
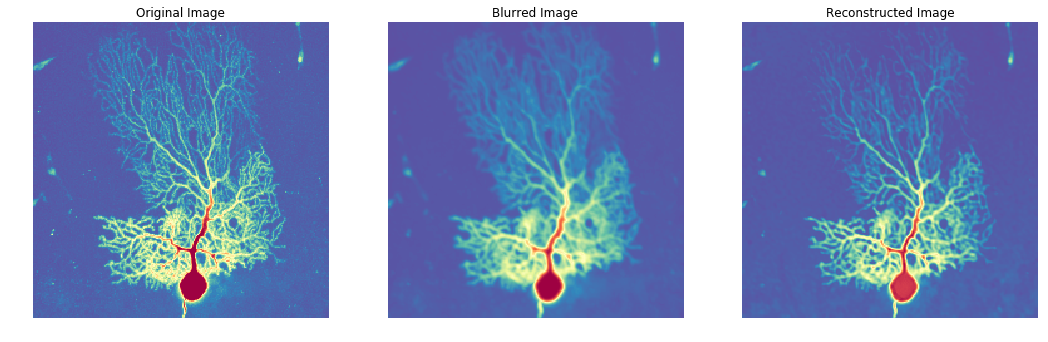
Here is a basic example demonstrating how Flowdec can be used in a single 3D image deconvolution:
See full example notebook here
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from skimage import exposure
from scipy import ndimage, signal
from flowdec import data as fd_data
from flowdec import restoration as fd_restoration
# Load "Purkinje Neuron" dataset downsampled from 200x1024x1024 to 50x256x256
# See: http://www.cellimagelibrary.org/images/CCDB_2
actual = fd_data.neuron_25pct().data
# actual.shape = (50, 256, 256)
# Create a gaussian kernel that will be used to blur the original acquisition
kernel = np.zeros_like(actual)
for offset in [0, 1]:
kernel[tuple((np.array(kernel.shape) - offset) // 2)] = 1
kernel = ndimage.gaussian_filter(kernel, sigma=1.)
# kernel.shape = (50, 256, 256)
# Convolve the original image with our fake PSF
data = signal.fftconvolve(actual, kernel, mode='same')
# data.shape = (50, 256, 256)
# Run the deconvolution process and note that deconvolution initialization is best kept separate from
# execution since the "initialize" operation corresponds to creating a TensorFlow graph, which is a
# relatively expensive operation and should not be repeated across multiple executions
algo = fd_restoration.RichardsonLucyDeconvolver(data.ndim).initialize()
res = algo.run(fd_data.Acquisition(data=data, kernel=kernel), niter=30).data
fig, axs = plt.subplots(1, 3)
axs = axs.ravel()
fig.set_size_inches(18, 12)
center = tuple([slice(None), slice(10, -10), slice(10, -10)])
titles = ['Original Image', 'Blurred Image', 'Reconstructed Image']
for i, d in enumerate([actual, data, res]):
img = exposure.adjust_gamma(d[center].max(axis=0), gamma=.2)
axs[i].imshow(img, cmap='Spectral_r')
axs[i].set_title(titles[i])
axs[i].axis('off')As a more realistic use case, here is an example showing how a point spread function configuration can be used in a headless deconvolution:
See full deconvolution script here
# Generate a configuration file containing PSF parameters (see flowdec.psf module for more details)
echo '{"na": 0.75, "wavelength": 0.425, "size_z": 32, "size_x": 64, "size_y": 64}' > /tmp/psf.json
# Invoke deconvolution script with the above PSF configuration and an input dataset to deconvolve.
# If flowdec has been installed, you may run the “deconvolution” command.
python examples/scripts/deconvolution.py \
--data-path=flowdec/datasets/bars-25pct/data.tif \
--psf-config-path=/tmp/psf.json \
--output-path=/tmp/result.tif \
--n-iter=25 --log-level=DEBUG
> DEBUG:Loaded data with shape (32, 64, 64) and psf with shape (32, 64, 64)
> INFO:Beginning deconvolution of data file "flowdec/datasets/bars-25pct/data.tif"
> INFO:Deconvolution complete (in 7.427 seconds)
> INFO:Result saved to "/tmp/result.tif"- Neuron - Deconvolution of a natural 3D image with synthetic point spread function
- C. Elegans - Deconvolution of 712x672x104 acquisition for 3 separate channels
- Astronaut - Dealing with artifacts in deconvolved images
- Monitoring Iterations - Tracking deconvolution progress by visualizing results at each iteration
- Hollow Bars - Deconvolution of downsampled 64x64x32 synthetic volume (as a CPU-friendly example)
- Hollow Bars GPU Benchmarking - Testing running times on full 256x256x128 volume with GPU-enabled system
- DeconvolutionLab2 Comparison - Comparing execution times between DeconvolutionLab2 and Flowdec
- Graph Export - Defining and exporting TensorFlow graphs
- Command Line Interface - CLI for executing single deconvolutions with either a pre-defined or dynamically generated point spread function
- Multi-GPU Example - Prototype example for how to be able to execute deconvolution against multiple GPUs in parallel (not tested yet -- waiting for the use case to come up though it is very likely possible to do)
The project can be installed, ideally in a python 3.6 environment (though it should work in 3.5 too), by running:
pip install flowdec[tf_gpu]The previous command will install flowdec, but also ensure that tensorflow
is installed with GPU support. For test purposes, you may have the non-GPU
enabled version of tensorflow installed by running:
pip install flowdec[tf]If neither [tf] or [tf_gpu] are specified, tensorflow installation is left
as an externally managed prerequisite.
Alternatively, the project could be installed from source by doing the following:
git clone https://github.com/hammerlab/flowdec.git
cd flowdec/python
pip install -e .A local docker image can be built by running:
cd flowdec # Note: not flowdec/docker, just cd flowdec
docker build --no-cache -t flowdec -f docker/Dockerfile .
# If on a system that supports nvidia-docker, the GPU-enabled version can be built instead via:
# nvidia-docker build --no-cache -t flowdec -f docker/Dockerfile.gpu .The image can then be run using:
# Run in foreground (port mapping is host:container if 8888 is already taken)
docker run -ti -p 8888:8888 flowdec
# Run in background
docker run -td -p 8888:8888 --name flowdec flowdec
docker exec -it flowdec /bin/bash # Connect The Flowdec dockerfile extends the TensorFlow DockerHub Images so its usage is similar where running it in the foreground automatically starts jupyter notebook and prints a link to connect to it via a browser on the host system.
The previous image is built from the current master branch of github.com/hammerlab/flowdec.git. To build an image using your local copy of the source instead, you can use this command:
docker build --no-cache -t flowdec -f docker/Dockerfile.devel .You may want to combine this with a bind mount of your local source tree into the running container. This setup will let you make edits to the source and have them immediately take effect in the running container.
LOCAL_SRC=$(pwd)
DEST_SRC=/repos/flowdec
docker run -ti -p 8888:8888 -v ${LOCAL_SRC}:${DEST_SRC} flowdecBy in large, the purpose of this project is to attain near equivalence with a subset of the functionality provided by both DeconvolutionLab2 and PSFGenerator via much faster implementations.
To validate this much has been accomplished, there are two notebooks in the python/validation folder demonstrating the following:
- Deconvolution Validation - This notebook aggregates results from Flowdec and DeconvolutionLab2 applied to several reference datasets and verifies that deconvolved volumes are very nearly identical
- PSF Generation Validation - This notebook aggregates results from Flowdec and PSFGenerator used to generate PSFs from a variety of different configurations and evaluates their similarity (which is also very high)
Thanks to Kyle Douglass for explaining some of the finer aspects of this Python Gibson-Lanni PSF generator, Jizhou Li for helping to better understand that diffraction model, Hadrien Mary for giving great context on the state of open-source deconvolution libraries, and Brian Northan for lending great advice/context on library performance, blind deconvolution and how point spread functions work in general.
- [1] D. Sage, L. Donati, F. Soulez, D. Fortun, G. Schmit, A. Seitz, R. Guiet, C. Vonesch, M. Unser
DeconvolutionLab2: An Open-Source Software for Deconvolution Microscopy
Methods - Image Processing for Biologists, 115, 2017. - [2] J. Li, F. Xue and T. Blu
Fast and accurate three-dimensional point spread function computation for fluorescence microscopy
J. Opt. Soc. Am. A, vol. 34, no. 6, pp. 1029-1034, 2017. - [3] Brandner, D. and Withers, G.
The Cell Image Library, CIL: 10106, 10107, and 10108.
Available at http://www.cellimagelibrary.org. Accessed December 08, 2010.