This is a demonstration repository for The Pitt Challenge project about medical image compression.
Health-care providers have to store a rapid growing collection of patients' medical records. And medical images occupies a big portion of the total size. In addition, The recent popularity of blockchain raises probabilty to serve medical records through similar fashion. However, blockchain as database works clumpsily, and thus is very sensitive to data size. Our team works to compress DICOM image files to a much smaller size and tries to propose a feasible way to serve on blockchain.
There are numerous image taken in the hospital everyday. But since image are usually taken about certain part of human body, great amount of similar images are produced. The core of our project use this property to compress data: encode images as predictions from a similar one.
This way, a set of similar images can be stored as a chain object contains the first original image and following predictions. Then, the original images can be reconstructed by performing such predictions stored in the chain object.
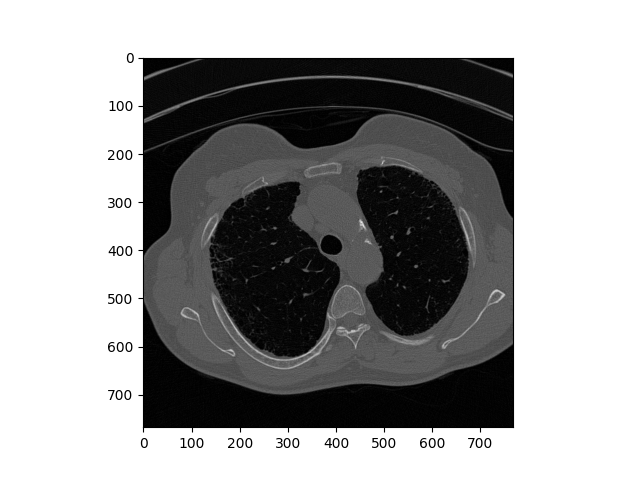
Original DICOM Image
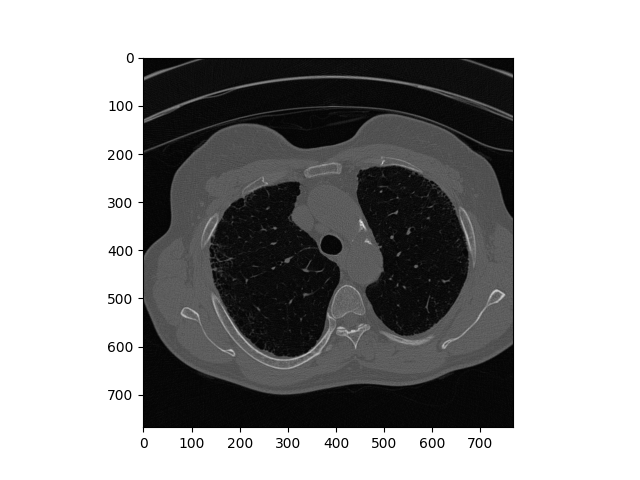
Reconstructed DICOM Image
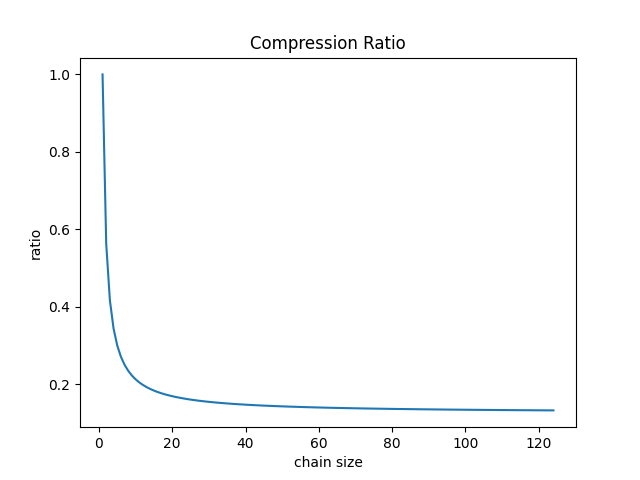
Above is the comparison of the original image and reconstructed one. This demo works on eight similar dcm images and the comparison is about the eighth image on chain. Overtly, the reconstructed image retains detailed feature and has no visual difference with the original one. In theory, larger chain size provides better compression ratio. It is worth to note that image could be not as precise as original when the chain grows too big. One naive solution is to limit the chain size.
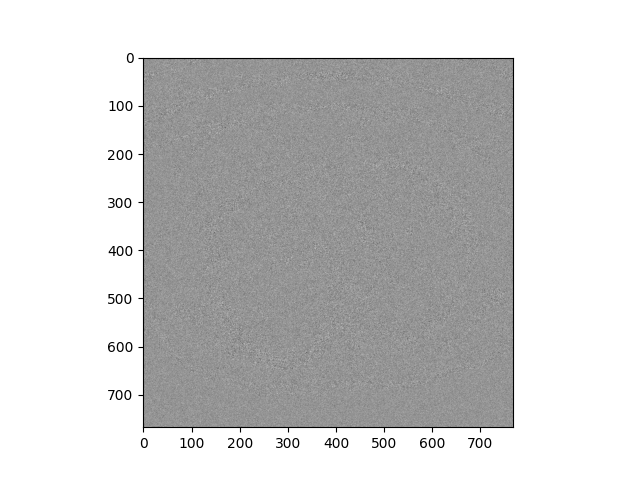
As mentioned in the previous section, the reconstructed image is not lossless. With our setup for demonstration(using lossy parameters on encoder), the difference is still neglectable -- about 3% difference at maximum. Following image is an visualization of this difference; obviously, it does not blur the detail feature of the original image.
difference between original and reconstructed images
On top of that, we found that the difference of features can surely be represented by a sparse matrix, i.e. we are able to compress the difference between two consequent images with square root of original image size, e.g. storing with Yale format.
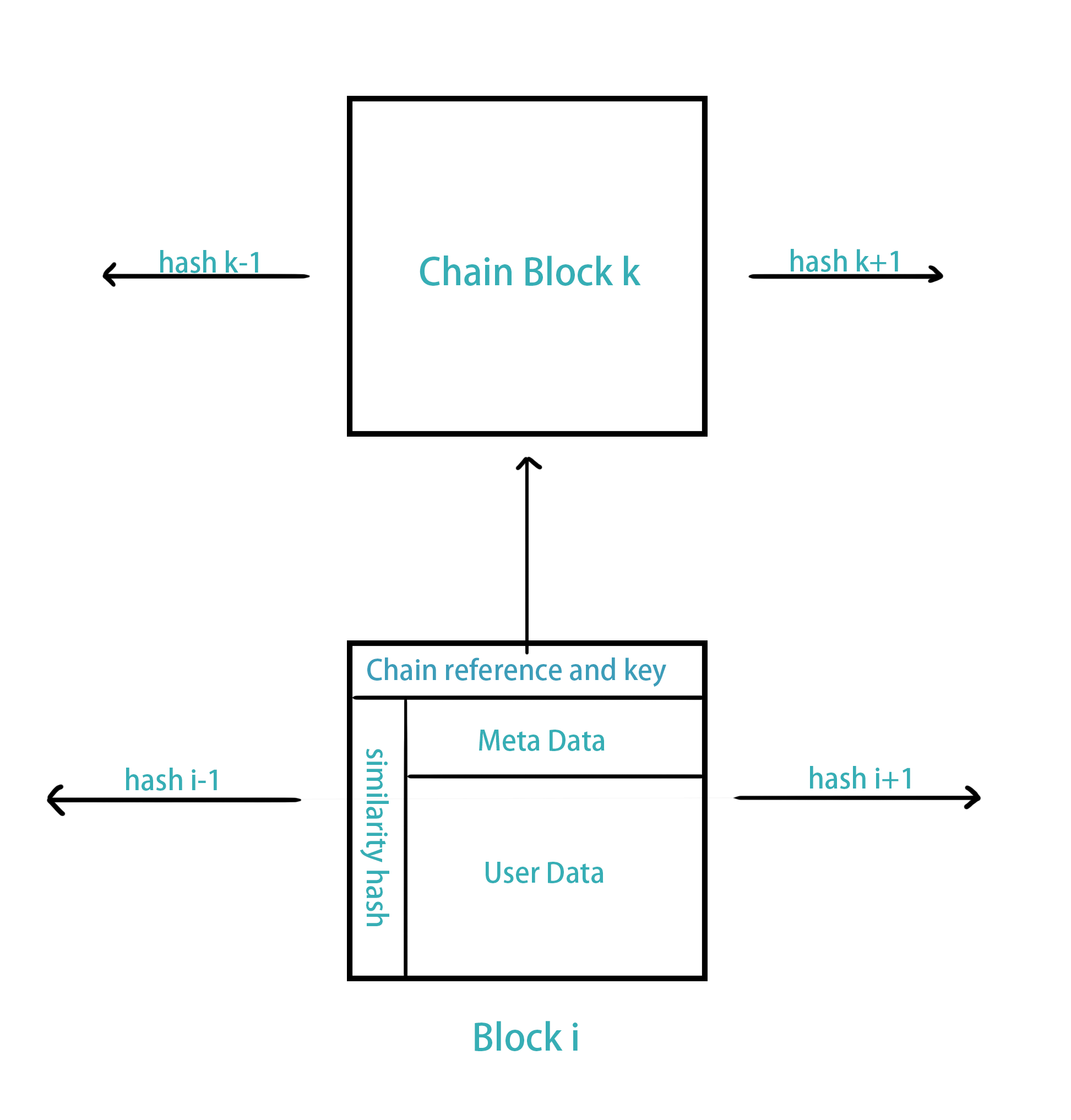
It is feasible to implement this discussed method on blockchain. For instance, we may set up two blockchains. One saves the integrated chain objects, and another chain saves similarity hash of image data, meta data of the DICOM file, other user data, and a reference to the other blockchain the chain object it depends on with a key to fetch its private image. To save a new image, the algorithm fetch blocks with adjacent similarity hash and the problem reduces to find the most efficient way to encode. This is actually an travelling salesman problem(to find the shortest path traversing all nodes), and indeed it can be solved, e.g. using simulated annealing. The other direction is rather simple. If a patient or a health-care provider need the DICOM image file, they simply use the patient's private key to ask the algorithm communicate to the other blockchain, and then the blockchain with chain object data will decode one's image using the key provided and send back the reconstructed image.
Blockchain Configuration
With this configuration, not only the data storage is saved on health-care providers' side, but also enables a secured, portable, lightweight storage for everyone's medical records.
This implementation demonstrates the compressing and reconstructing process of similar DICOM images.
- Tested only on Windows10
- MinGW
- Python-3
- vapoursynth
- x265
Above tools are used only for the sake of simplicity. In real case, for example, we may use a specific encoder designed for such task, instead of x265, to achieve better result.
It is easy to compile this demo source. We use make command come with MingW:
% cd <repository path>
% make
Now, there will be a build directory in the top directory, in which there are executable files needed for demonstration.
Lastly, you have to manually move the libvsmat.dll file to vapoursynth's plugin directory(you will need administrator's permission to do so). Alternatively, you can type:
% mv build/libvsmat.dll <vapoursynth plugin path>
First, you move several similar DICOM images to the build directory. If you do not have, there are eight files in the sample directory you may use. Then the command:
% .\encode.exe
compresses the images in the build directory as the methodology section described, and compressed data are presented in the encoded subdirectory.
To extract the last image (for the sake of time, this demo only support extraction of last DICOM file using following command. But it is actually easy to do it for every images. in fact, you may tweak the source code just a little bit to achieve such result), you type as following:
% .\decode.exe <basename of your original DICOM file, e.g. 17 for 17.dcm>
This will reconstruct your last dicom file to the decoded subdirectory.
To see the images, you may you following python command:
% py dcmshow.py <your DICOM file>
Besides, following python script is useful to visualize the difference between two dicom images"
% py .\dcmshow_diff.py <DICOM file1> <DICOM file2>
This command will also output the maximum difference between two images. In our case, we have following:
% py .\dcmshow_diff.py .\17.dcm .\decoded\17.dcm
maximum percent difference: 0.028788881535407016
If you follow above instructions, the resulted files in build directory should be like as below:
build
├── [1.1M] 10.dcm
├── [1.1M] 11.dcm
├── [1.1M] 12.dcm
├── [1.1M] 13.dcm
├── [1.1M] 14.dcm
├── [1.1M] 15.dcm
├── [1.1M] 16.dcm
├── [1.1M] 17.dcm
├── [1.4K] dcmrc.py
├── [ 197] dcmshow.py
├── [ 91K] decmat.exe
├── [ 90K] decode.exe
├── [1.1M] decoded
│ └── [1.1M] 17.dcm
├── [ 54K] encode.exe
├── [1.0K] encode.vpy
├── [1.5M] encoded
│ ├── [ 4] 10.max
│ ├── [1.3K] 10.meta
│ ├── [ 4] 11.max
│ ├── [1.3K] 11.meta
│ ├── [ 4] 12.max
│ ├── [1.3K] 12.meta
│ ├── [ 4] 13.max
│ ├── [1.3K] 13.meta
│ ├── [ 4] 14.max
│ ├── [1.3K] 14.meta
│ ├── [ 4] 15.max
│ ├── [1.3K] 15.meta
│ ├── [ 4] 16.max
│ ├── [1.3K] 16.meta
│ ├── [ 4] 17.max
│ ├── [1.3K] 17.meta
│ ├── [1.4M] chain.hevc
│ └── [ 124] chain.hevc.ffindex
├── [ 309] image.vpy
├── [ 64K] libvsmat.dll
├── [ 212] plot.py
├── [ 611] save_meta.py
└── [ 576] write_mat.py
12M used in 2 directories, 38 files
Team: BadPineapple