PKNER| Setup | Download data and models | Inference | Citing
This repository contains code to perform Named Entity Recognition of Pharmacokinetic Parameters in the scientific literature.
-
Create and activate a virtual environment with
python 3.8.12installed -
Install this repo to get started:
git clone https://github.com/PKPDAI/PKNER
cd PKNER
pip install -e .sh scripts/download_annotations.sh
sh scripts/download_pretrained_biobert_pkner.shTo use NER for PK parameters with spaCy make sure scispaCy is installed (pip install scispacy). Then install the NER package for PK parameters through:
pip install https://pkannotations.blob.core.windows.net/nerdata/trained_models/en_pk_ner-0.0.0.tar.gz
You can use the model through:
import spacy
nlp = spacy.load("en_pk_ner")
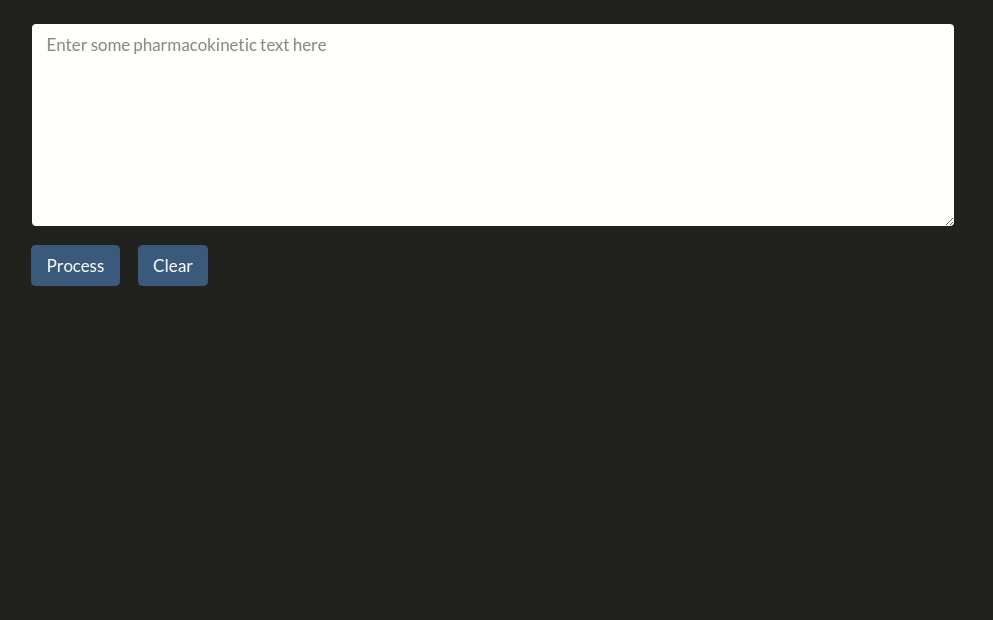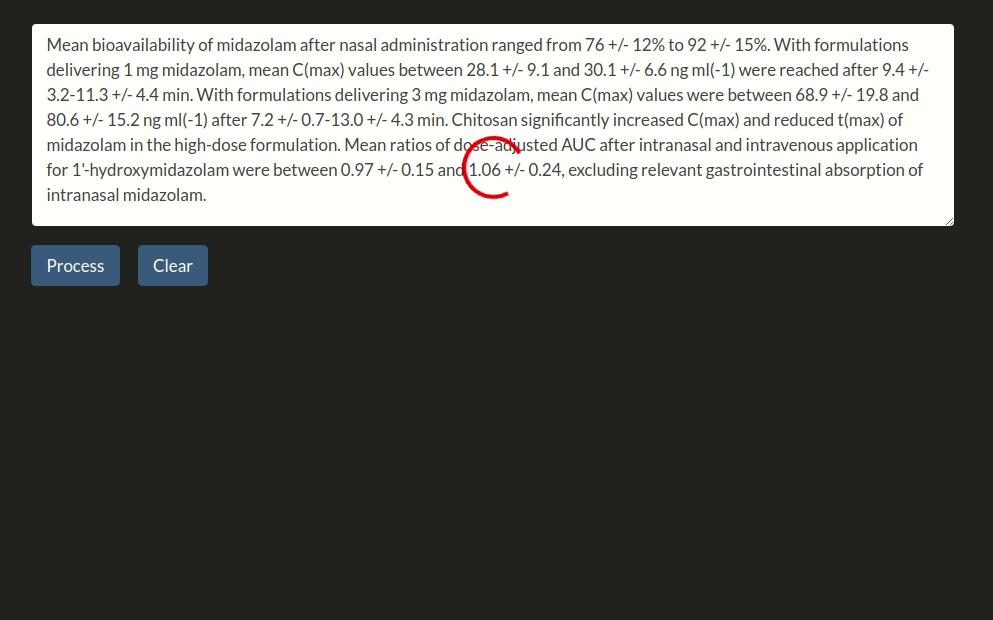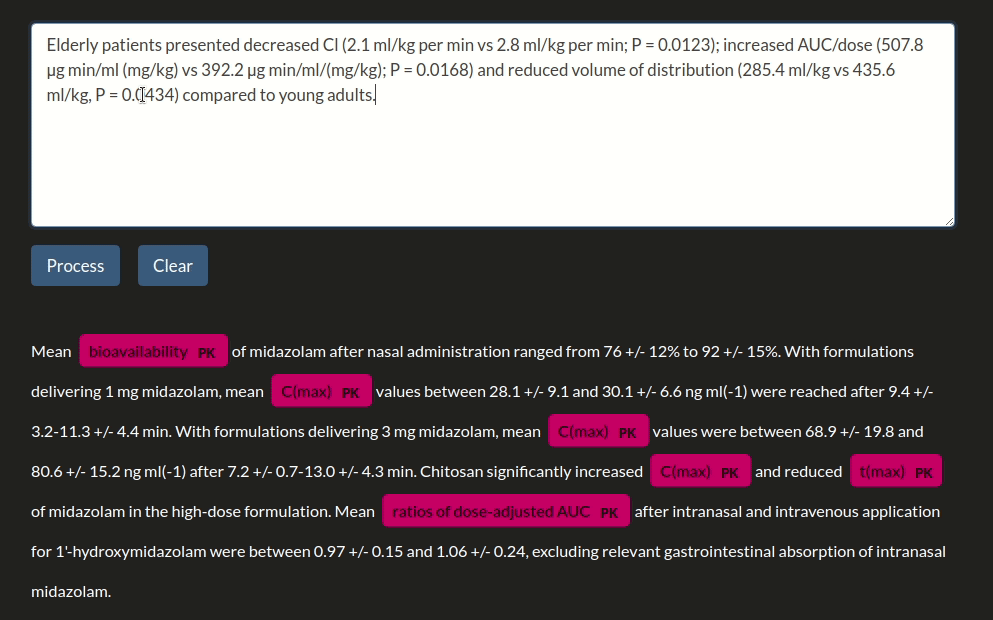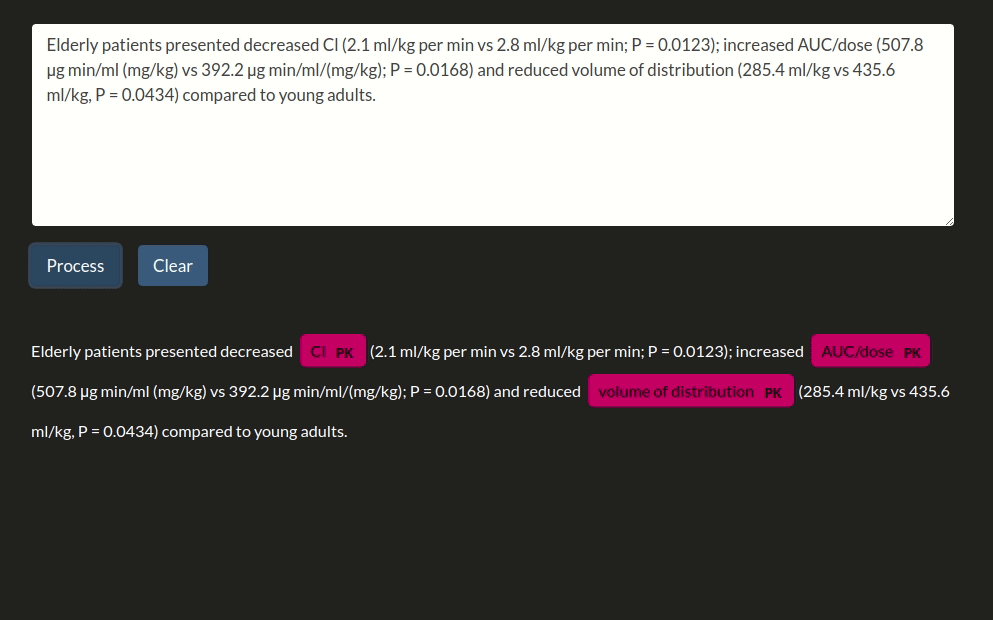
doc = nlp("Parameter estimations for a subject of 34kg indicated values of midazolam clearance of 34.7l·h-1, a central volume of distribution of 27.9l and a peripheral volume of distribution of 413l.")
for ent in doc.ents:
print(ent)
#>>> clearance
#>>> central volume of distibution
#>>> peripheral volume of distributionpython scripts/evaluate_bert.py \
--model-checkpoint checkpoints/biobert-ner-trained.ckpt \
--predict-file-path data/test.jsonl \
--display-errors \
--batch-size 256 \
--gpu \
--n-workers 12@article{hernandez2024named,
title={Named Entity Recognition of Pharmacokinetic parameters in the scientific literature},
author={Hernandez, Ferran Gonzalez and Nguyen, Quang and Smith, Victoria C and Cordero, Jose Antonio and Ballester, Maria Rosa and Duran, Marius and Sole, Albert and Chotsiri, Palang and Wattanakul, Thanaporn and Mundin, Gill and others},
journal={bioRxiv},
pages={2024--02},
year={2024},
publisher={Cold Spring Harbor Laboratory}
}