Alrighty, let's get into it. The goal in this project is to go from the chaotic assortment of scripts and independent work
in the 1_starting_point directory, into something that looks like a much nicer DS pipeline.
For windows users, I'd recommend first running conda install sqlalchemy==1.3.13 so you don't have issues with wheels building.
pip install -r requirements.txt
Within the starting_point directory, I've tried to simulate the somewhat chaotic and unstructured way that some data science projects
begin. A combination of different people sharing parts of the workflow, without a consistent
structure or development route set in place.
1_starting_point
- make_processed_data.ipynb
an initial EDA notebook that took raw data, made plots and produced processed data
- model_linreg.ipynb
a quick and dirty linear regression model to the data
- model_keras.ipynb
an untuned and small neural net to do regression to the data
- germany.csv
The processed data generated from the first notebook
- report
Directory containing a weekly report (and its creation script) that should go out to clients
- dashboard
Directory containing a Dash dashboard to provide live insight into the data
The first goal will be to take the models and improve the workflow on training them, recording their hyperparameters, logging their output artifacts and metrics.
Then once we have the models logged, it is easier to pick a model for production, so we'll cover staging models and deploying them locally.
Finally, we'll turn the data processing and report into a multi stage workflow, and hook the dashboard up to the production model.
First thing we want to do is to start a tracking server. Instead of just pointing this at the file system and hoping for the best, lets put in a tiny bit of effort now so that it better emulates a remote tracking server like you might actually end up using.
With the tracking, we need a place to put information, and a place to put artifacts. The latter is normally a google cloud, Azure blob storage, Amazon S3 bucket, etc. For now, we'll just have a super simple local directory. Here is what I would run:
mlflow server --backend-store-uri sqlite:///mlruns.db --default-artifact-root file:/home/samreay/mlruns
If you have any issues, you can run the smaller version (which uses the file system for simplicity but doesnt support model registry, so it won't work with the airflow part later)
mlflow ui
Optional step: Now one thing we want to do is make sure that this is the server we use for the entire workshop, so lets set a single environment variable. In yet another terminal window that I opened in the DSGOPipeline folder, I will now set:
export MLFLOW_TRACKING_URI=http://127.0.0.1:5000
which is just the location of the server we launched. Note that because environment variables are an easy thing to get wrong or for people to do incorrectly, I also set it explicitly in all the code, so you don't need to do the above. But ideally you wouldn't set anything in the code!
And that's it! Now we jump into the code by starting up jupyter notebooks back in the project directory (wherever you clone or downloaded this repo to):
jupyter notebook
So you'll have one terminal window (in your home directory) running the mlflow tracking server. And you'll have another terminal window in the project directory running jupyter notebooks. Keep both of these terminals up and running for the whole workshop.
Let's first run through the 1_starting_point folder notebooks, so we can see what's going on. And then we'll want to go through each
of those notebooks and convert them to make use of mlflow. You can see the changes in the 2_ml_tracking folder.
- Starting with the linear regression, we see how simple it is to log metrics and the model.
- With the RFRegressor, see how we can also log model parameters. And then see that you have multiple runs in a single execution, if you really want!
- Then with Keras, we make use of autolog, which some libraries have support for (sklearn is not one of them yet, because there are so many different models in sklearn!)
At any point during the above, you can open up the tracking server (in your browser go to http://127.0.0.1:5000) and you can see the metrics, parameters, models and artifacts under the "predicting_solar_wind" experiment. If you try and run them all at the same time, the SQLite DB is going to have an issue with you as it only supports sequential writes.
This tracking raises a few questions:
- How can we compare multiple models ourselves, outside of the UI interface? (Note that this will probably be built into mlflow at some point, and atm tools like Neptune can do it for you, for now as a manual way, see the compare_runs notebook).
- How can someone reproduce the exact run? (mlflow projects)
- How can we pick a model to serve? (mlflow model repository)
Check out the compare_runs.ipynb notebook in the 2_ml_tracking folder.
If we want hands on review, this is super easy. Simply go to the experiment, find the "best performing" algorithm that you want to be staged, and then click on it. We might decide, for example, that the run with the minimum MAE is the one we want, so we can sort the runs on MAE and pick the lowest one, clicking into it. Under artifacts, click on model, and you'll see Register Model. So I made a new one, called BestModel, which is now registered.
I can then transition the model into either staging or production, by clicking on it again. Let's move it into production. But now, how can we use it?
Note: access tracked models by going to localhost:5000/#/models
For an example fetching this model via the API, see the get_prod_model.ipynb.
We can also serve it locally (or deploy it to SageMaker, AzureML, Spark, etc). Here is the command to do it locally (note the source of the model, which is printed out when you run the notebook above, and you'll have to change this to your location)
mlflow models serve -m file:///home/samreay/mlruns/1/f0fd5630bb234ba89295d7d34b47254b/artifacts/model -p 8003 --no-conda
which will expose an endpoint you can use directly to run model predictions without having to run literally any code on your own. If you run the above line (in a new terminal window) and then (in yet another new terminal window) run this curl, you should get back predictions:
curl -d '{"columns": ["windspeed", "temperature", "rad_horizontal", "rad_diffuse"], "data": [[2, 20, 100, 100]]}' -H 'Content-Type: application/json; format=pandas-split' -X POST localhost:8003/invocations
Once we do have a model that is in production, we can use the code from get_prod_model.ipynb and incorportate that into our
weekly reports and dashboard, solving that issue completely.
import mlflow.pyfunc
from mlflow.tracking.client import MlflowClient
model_version = MlflowClient().get_latest_versions("ModelName", stages=["Production"])
prod_model = mlflow.pyfunc.load_model(model_version[0].source)Lets bundle up our linear regression model into a simple MLFlow project. A project is defined by the MLproject file, its super simple and can have multiple stages.
name: projectName
conda_env: conda.yaml # Or a docker container
entry_points:
main:
parameters:
some_path: path
some_param: {type: float, default: 1}
command: "python your_file.py {some_param} {some_path}"
validate:
command: "python some_other_file.py"First up, we can easily turn each notebook into an mlflow project, but that would take a while, so we can instead look only at the keras model.
To make the conda.yaml step easy,
don't ask conda for your current environment, just go to your tracked mlflow runs and it'll have a minimal version! But note it only tracks
what is used in the actual experiment after you call start_run, so you might need to add extra stuff in.
We'll wrap up the keras example (and not worry about pulling the dataframe as a path argument), so we can run (in another terminal window in the 3_ml_projects directory)
mlflow run keras_standalone --no-conda --experiment-name predicting_wind_solar. Note again we specify the experiment name and URI via command line / environment variable,
rather than defining it in our code. no-conda simply because I already have a working environment, and don't need a new one! If we were inside the directory, we could also run
mlflow run ., and of course this can all be done in code:
mlflow.run(project_dir, entry_point, parameters=parameters)At this point, we could commit the keras_standalone directory (with the conda.yaml, MLproject, etc) as its own git repo, and then invoke
mlflow run git@github.com/username/projectname.git --no-conda --experiment-name predicting_wind_solar, so you can have each step version controlled.
So we could use the MLProject formalism to package up everything in here, but this still leaves us with a bit of an issue, in that its still rather manual.
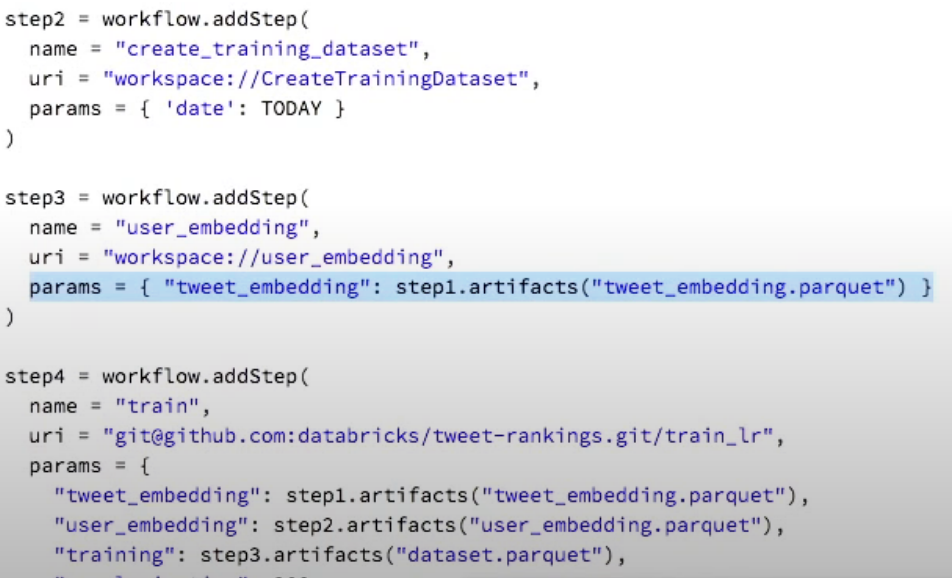
Whilst the mlflow team does have an example on cached multi-step workflows here, the example is still very manual and is back from 2018. They are currently developing a better automated system of keeping track of caching and having workflows defined as DAGs. They were teased during the 2019 keynote last year (https://www.youtube.com/watch?v=QJW_kkRWAUs, 28min), and I am hoping to hear good news on their development at the Spark AI summit that occurs the same week as this conference. So, fingers crossed.
Here is a screenshot from the keynote as to how (last year) they were coding up their DAGs:
Where you can see that inputs and outputs can be linked directly to the artifacts logged into the mlflow environment. I'm very keen for this.
In the meantime, there is either their multi-step workflow example as linked above, or you could investigate a more established technology, like Apache Airflow.
Now at this point I should state that airflow is a server tool, and installing this under windows is going to very likely cause you a massive headache.
If you are in windows, I'd recommend using the Ubunutu subsystem you can install (via the microsoft app store) to try and get around this, but please don't get caught up if you have installation difficulties.
At this point, let's simplify things and close down the mlflow servers we started up before so that we don't have as many things up in the air!
On terminal, this will install airflow, create a home directory for it, and set up an environment variable for you to use. The directory Ive specified below is the default. So Ill open a new terminal window (which I set to the home directory straight away).
cd ~
pip install apache-airflow
mkdir airflow
Note airflow is not part of the requirements.txt file, because it may not play nice with windows.
Start up the SQLite database, which will be good enough for a small example like this.
airflow initdb
And then we start the web server as well!
airflow webserver
You should now be able to launch http://localhost:8888 and see a bunch of empty examples. To define a new DAG, it goes
inside the $AIRFLOW_HOME/dags folder. And a DAG is just a python file defining tasks (aka operators) and how they connect.
You can see a list of possible operators here, but they include:
bash_operator: The task executes a command on bashpython_operator: The task executes some python function / commandpostgres_operator: The task executes some postgres query / commanddummy_operator: A task which does absolutely nothing (but can be used to group other tasks into a single "block")
And a ton more. So you may not have to create everything from scratch. Now I have a DAG in the 4_airflow/mlflow_dag.py folder, so I'm going to copy this into
airflows dags dir in a new terminal (note that the AIRFLOW_HOME should be re-exported, added to bashrc, or just ignore it because I made sure its in the default location anyway)
mkdir ~/airflow/dags
cp /github_location/4_airflow/* ~/airflow/dags
and then to have airflow pick it up, I'm going to start the scheduler from a new terminal in my home directory. (Note having the CSV file in the DAGs dir is very bad practise)
airflow scheduler
If you're keeping track, this is now four terminal windows we have open at once:
- mlflow tracking server (accessible at localhost:5000)
- jupyter notebook (accessible at localhost:8888)
- airflow webserver (accessible at localhost:8080)
- airflow scheduler (in the background)
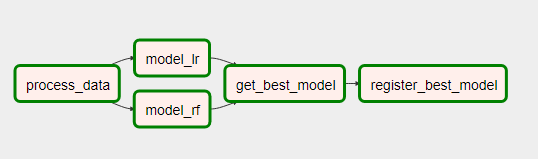
Back on topic! If we refresh our webserver page, you can see a new DAG has appeared! We can click on it and see each of the tasks in either the tree view or the graph view. And we can turn it on or off if we want it to execute as per the cron schedule configured in the DAG file.
We can test tasks piece by piece if want. To test just the process_data task, we can open a new terminal (in our home directory or wherever you want) and run:
airflow test DSGo process_data 2020-06-14T00:00:00.0
Back in the browser, if you refresh you can see that the process_data block has now changed outline. I hope its green!
We can kick the whole thing off either by running airflow trigger_dag DSGo or just clicking the UI button. It'll take a little while for each task to flow into another, don't worry if this takes a few minutes.
Before doing that, we should probably go through the code in mlflow_dag.py
Note you can embed ipython or debuggers as well if there are issues with a particular task and you are running on a task by task basis. See this link for details on debuggers.
Finally, what we'd ideally want to do is stage a best model for review each day, but for now the code just pushes models out to prod. Check out the registered model version (the URL is http://localhost:5000/#/models, hard to find in their current UI), you can see that the version in production will increment every time you run the DAG!
If we really wanted, we could generate the report (the jupyter notebook) using the new production model every time the DAG executes, which would provide some plots and predictions we could use to try and pick up any issues quickly (though ideally this would happen well before prod...). We could add this as something external, or just include it as another step in our DAG!
- You'd need to set up airflow, a backend data store and an ML tracking server properly.
- The little python operators in airflow can become bash operators which just execute MLProjects on arbitrary infrastructure. Because you're only passing back and forth run IDs, theres no worry about data flow.
- Better add some checks and validation on your production models.
To try and complete the circle, you can see in 5_evergreen_dashboard an updated version to utilise the production model. See the changes in components/elements.py on lines 35-43.
If you open a terminal window here, run python app.py and go to http://127.0.0.1:8050/ you can see it should be giving live predictions using a production model. Hurray! If there's time left now,
lets go through the dashboard code, because being able to slap together a dashboard like this in about 5 minutes is a super handy skill.