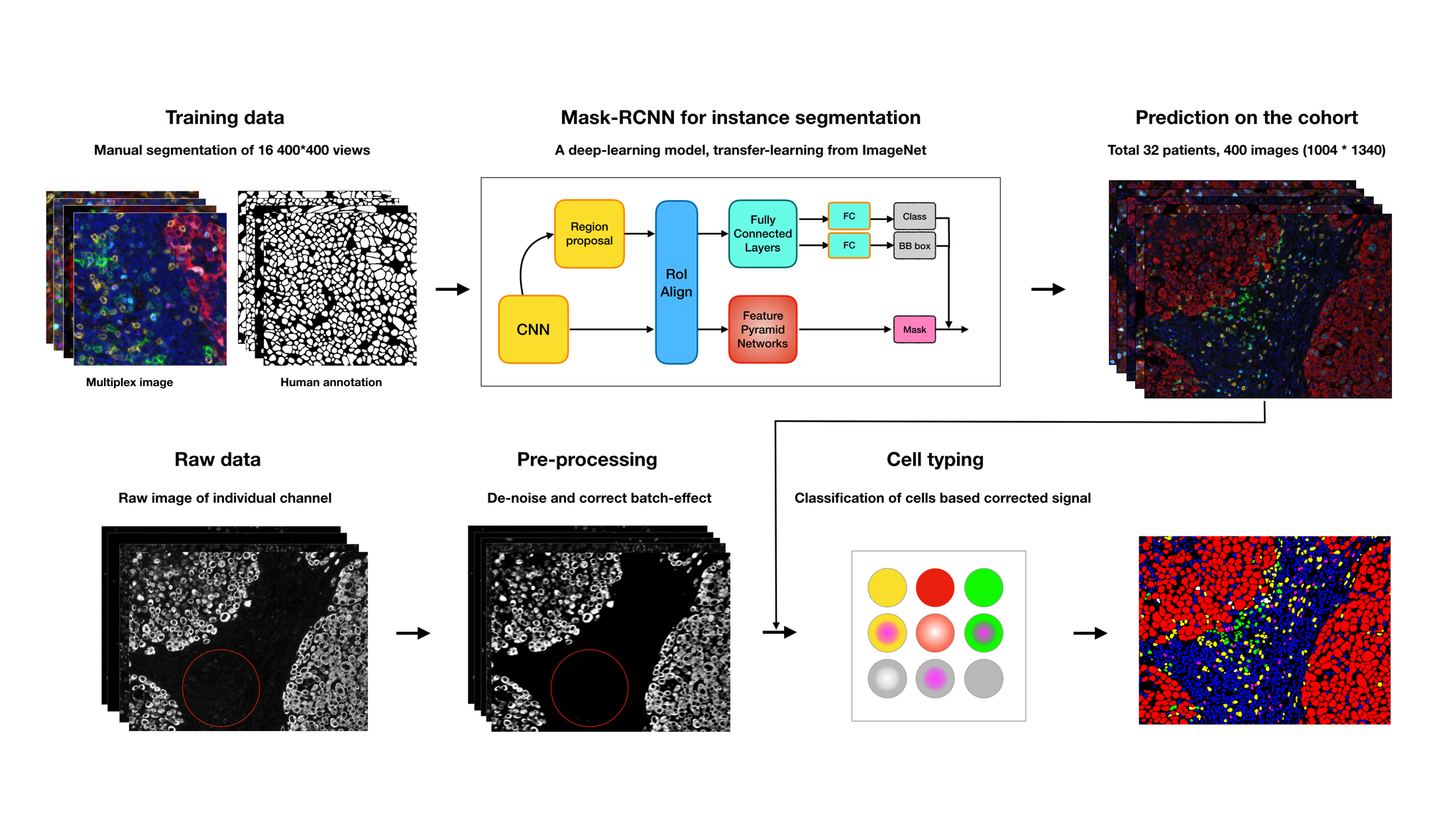
Multiplexed imaging data is providing dramatic opportunities to understand the tumor microenvironment, but there is an acute need for better analysis tools. Here, we provide a pipeline for multiplexed imaging quality control and processing. It contains three core steps:
- Preprocess raw images to remove undesired noise (introduced by technical sources) while retaining biological signal
- Perform segmentation to draw boundaries around individual cells, making it possible to discern morphology and which features, such as detected RNA or protein, belong to each cell.
- Extract cellular feature from images via segmentation and assign cell types to each cell.
To transform digital images into cell-level measurements, we have been applying, comparing and optimizing cutting-edge computer vision and machine learning techniques to each of the steps above. All code is written in Python.
-
Clone this repository
-
Create a new conda environment for this pipeline
-
conda env create -n davinci -f environment.yml source activate davinci python -m ipykernel install --user --name davinci -
Install this package
-
python setup.py install
-
Install the deep learning model for segmentation following the instruction on link
Step-by-step tutorial on the usage can be found in the following Jupiter notebooks.
More details can be found in the technical notes
- Train deep learning model on custom data Notebook
- Predict segmentation with pre-trained model Notebook
- Provide more pre-trained weights for segmentation using different marker panels
- The data in this repo are for demonstration only. Please do not use them for any other purposes.
This work is supported by
- The Alan and Sandra Gerry Metastasis and Tumor Ecosystems Center
- Human Tumor Atlas Network
- Parker Institute for Cancer Immunotherapy
He, K., G. Gkioxari, P. Dollár, and R. Girshick. 2017. “Mask R-CNN.” In 2017 IEEE International Conference on Computer Vision (ICCV), 2980–88. link