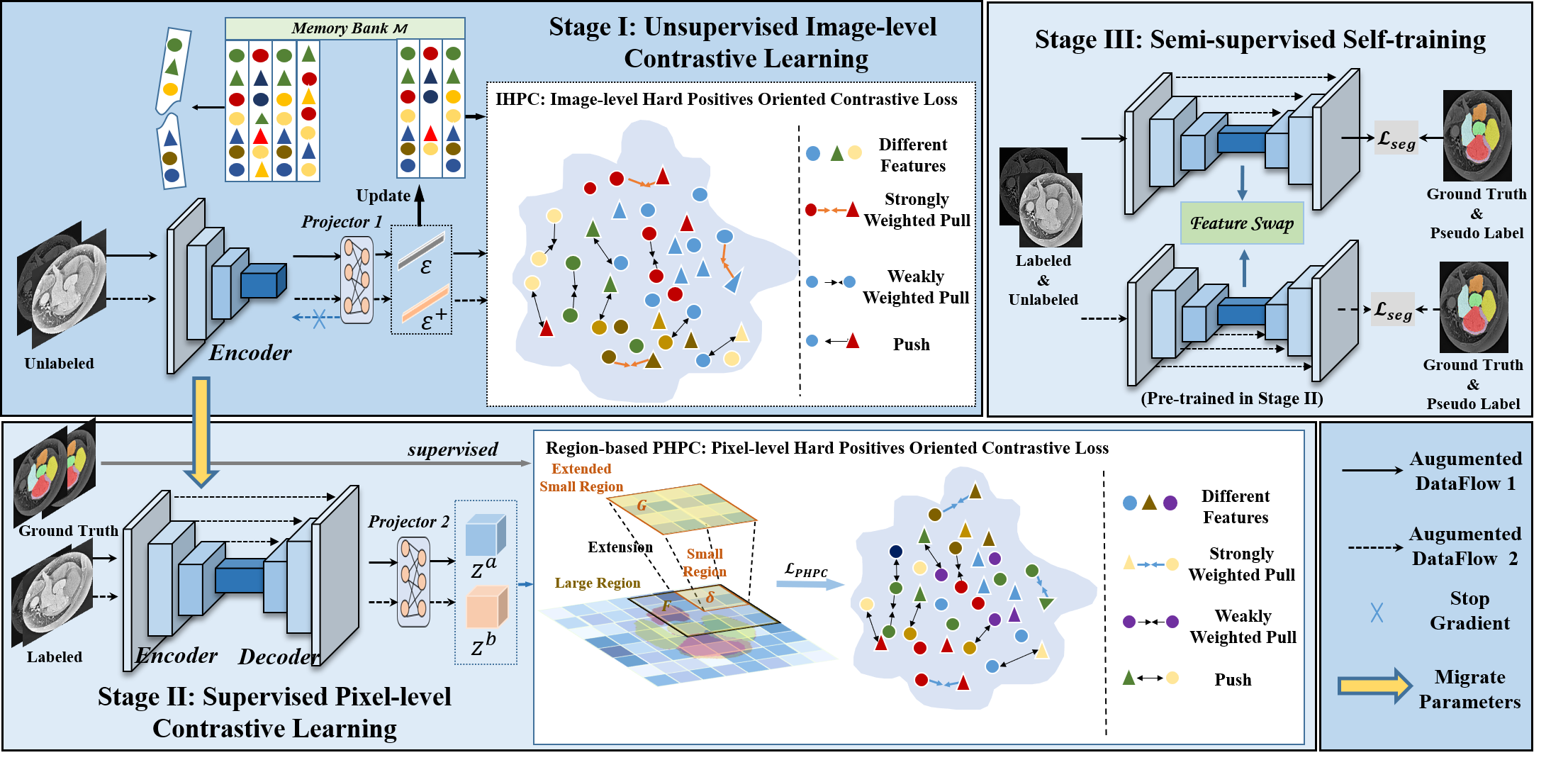
This is the pytorch implementation of paper "Semi-supervised Medical Image Segmentation via Hard Positives oriented Contrastive Learning".
Email addresses: tangcheng1@stu.scu.edu.cn & perperstudy@gmail.com (Joint First Authors) & wangyanscu@hotmail.com (Corresponding Author)
python=3.7.10
torch==1.8.1
torchvision=0.9.1
We will take the Hippocampus dataset as the example to illustrate how to do the preprocessing. Put the images .nii.gz files in ./data/Hippocampus/imgs folder and labels files in ./data/Hippocampus/labels.
cd dataset/prepare_dataset
python preprcocessing.py
python create_splits.py
Afterwards, the images and their respective labels will be combined and saved in a .npy file. The shape of the images will be normalized to match the target size.
To run the Stage I: Unsupervised Image-level Contrastive Learning,
bash run_simclr.sh
To run the Stage II: Supervised Pixel-level Contrastive Learning,
bash run_coseg.sh
To combine the above two pretraining, run run_simclr.sh first and the pretrained model will be saved at save/simclr/Hippocampus/ and set --pretrained_model_path ${the saved model path} in run_coseg.sh. Remember to load the saved pre-trained model.
To run the Stage III: (Semi-supervised Segmentation),
bash run_seg.sh
- In all of the aforementioned three files, "train_sample" denotes the percentage of labeled data to be utilized.