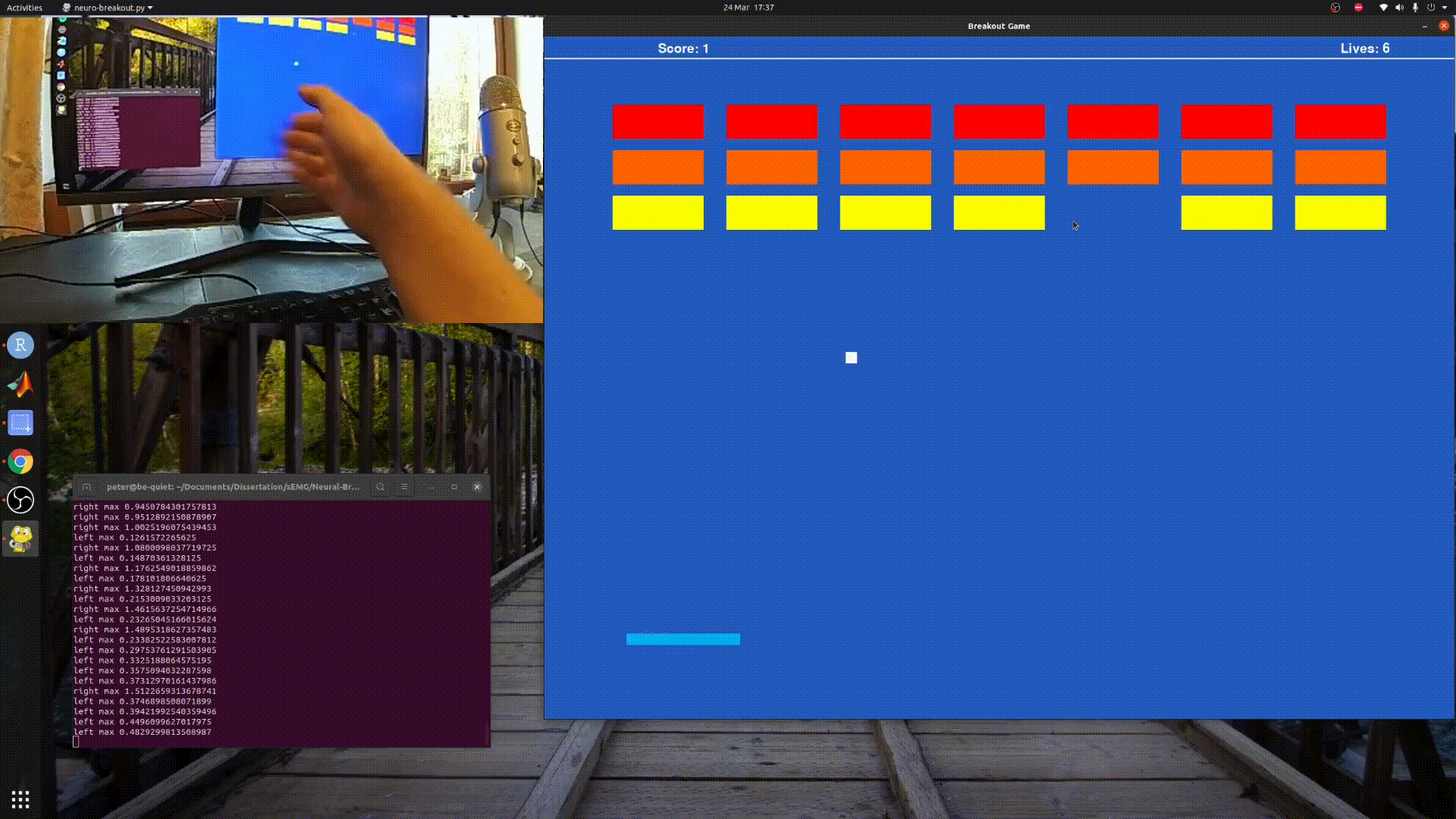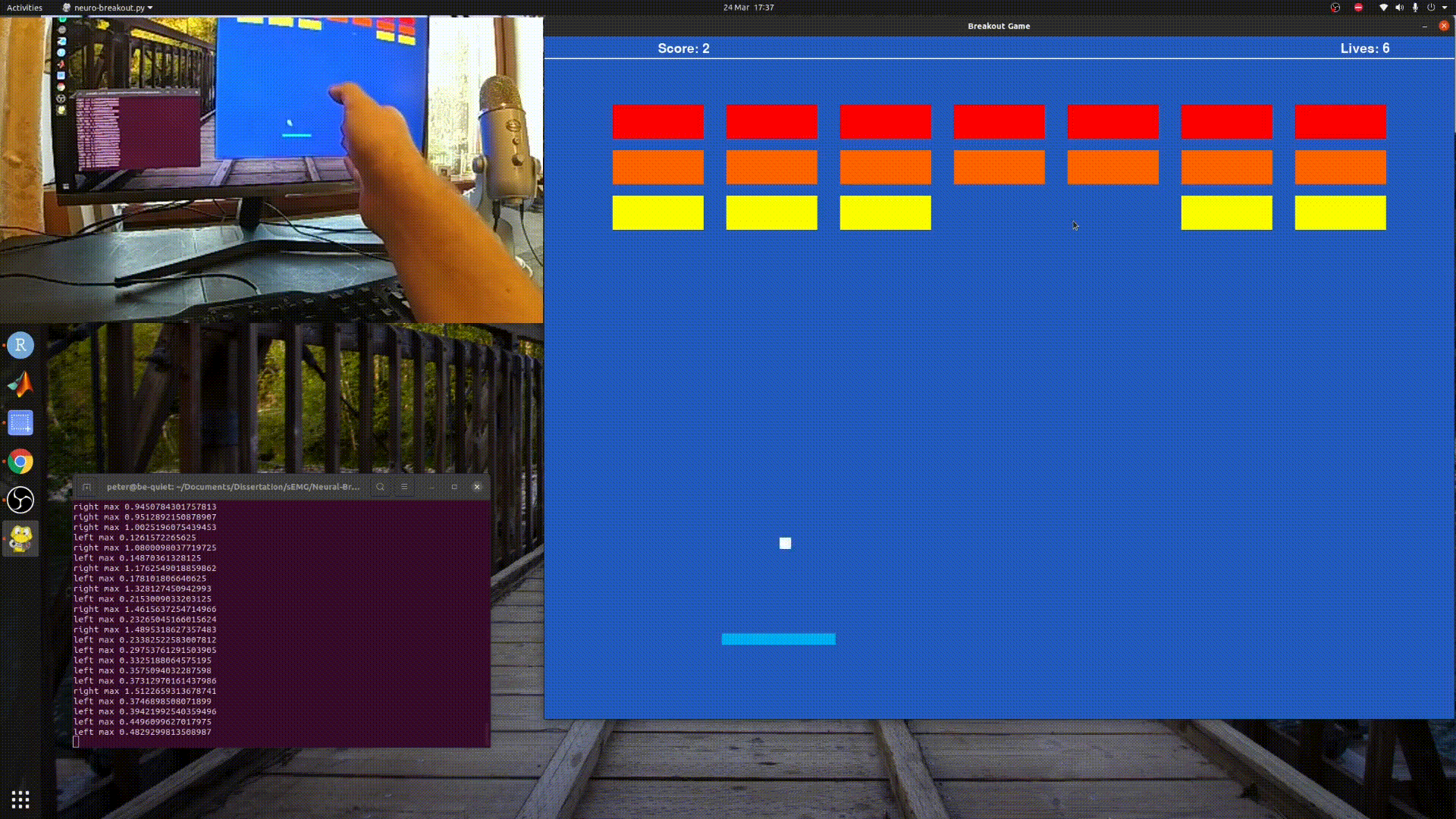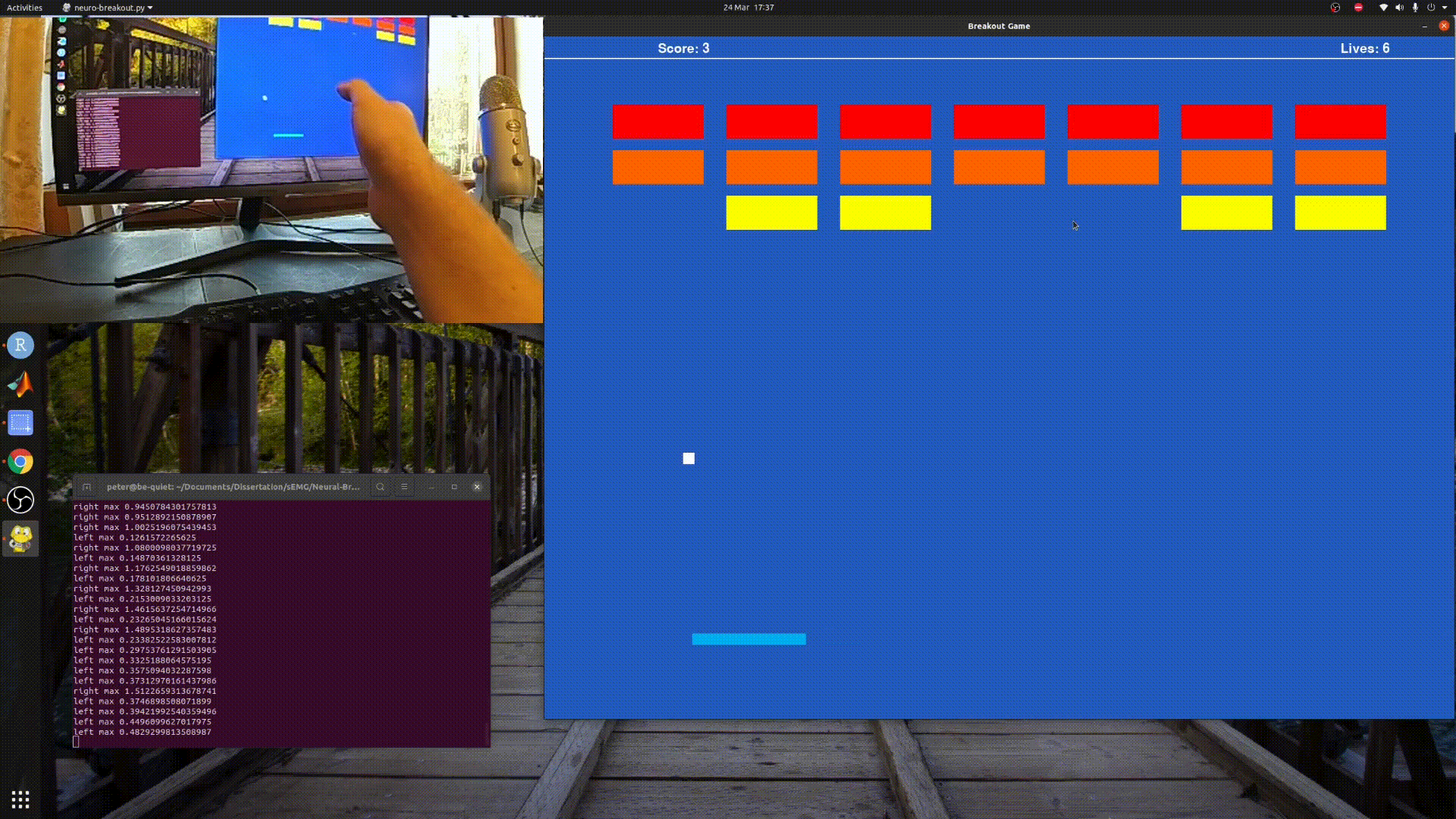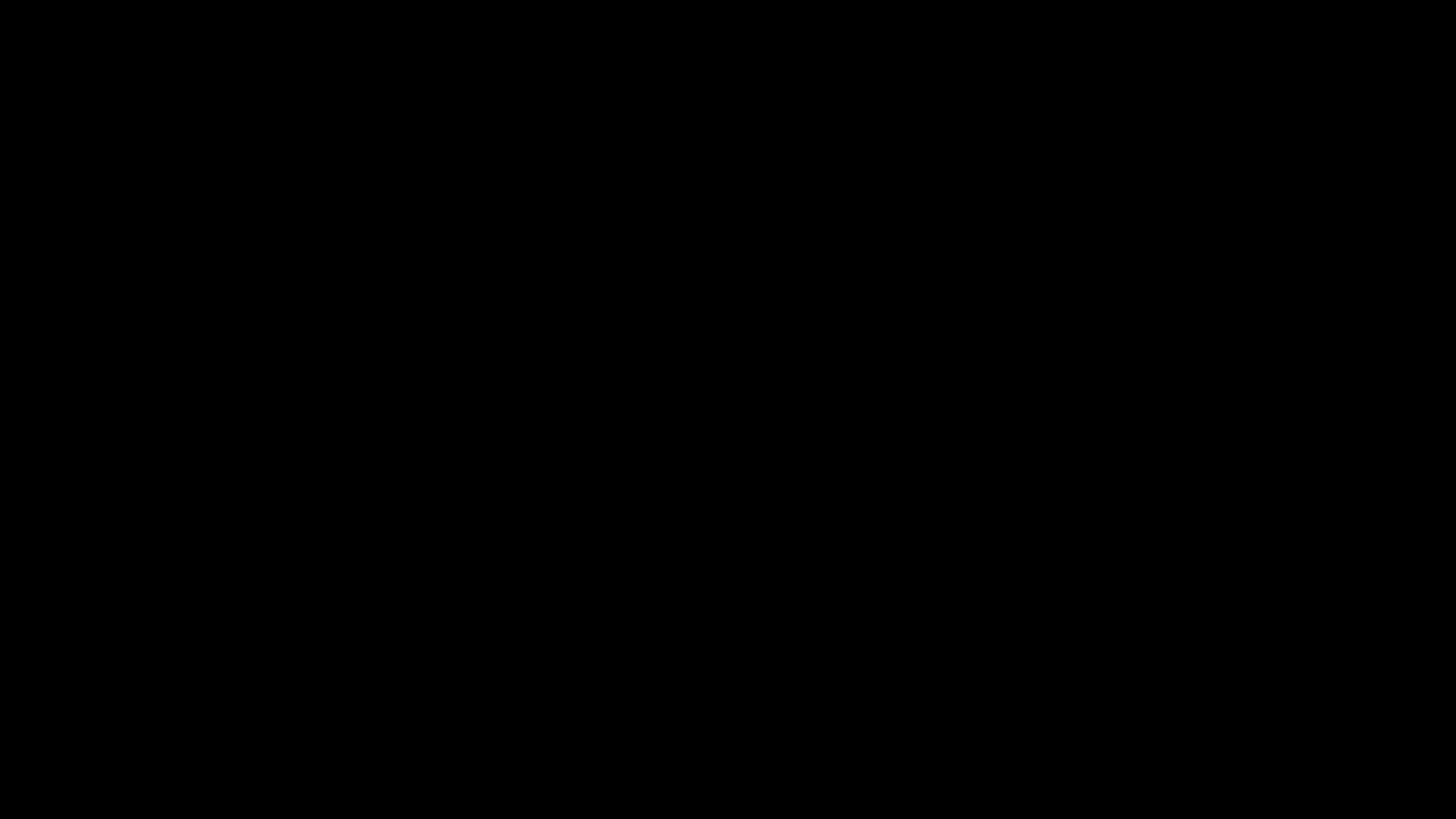
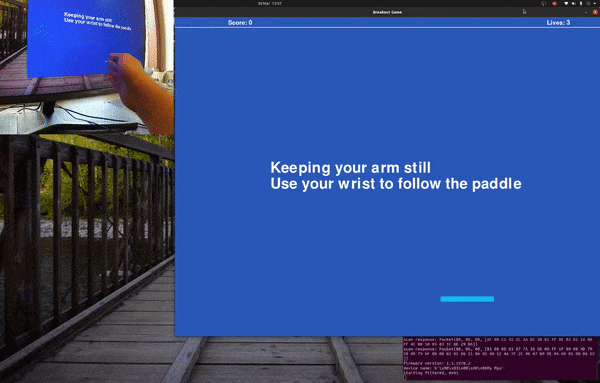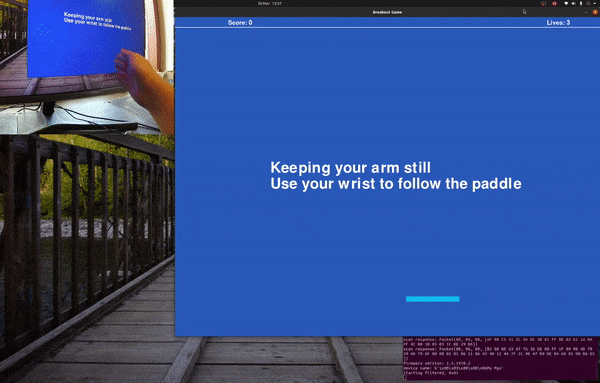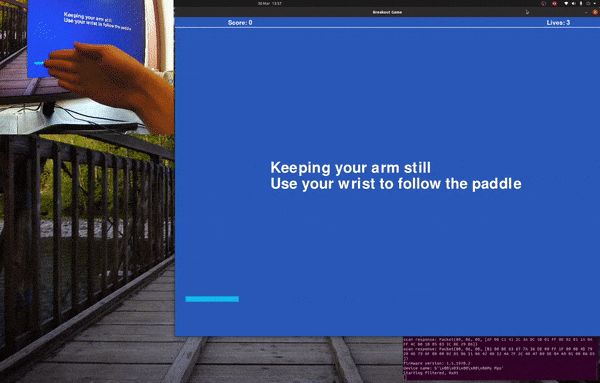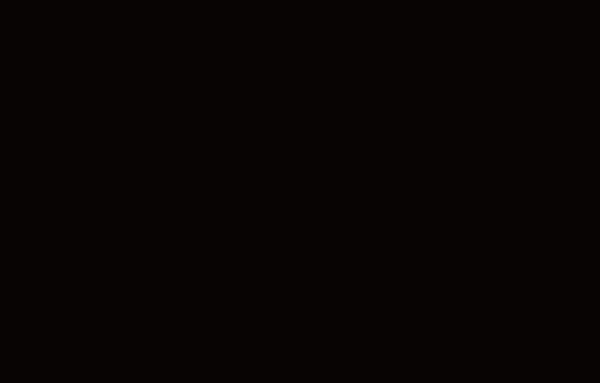
Playing Breakout with sEMG and Myo Armband.
pip install pyomyo==0.0.2
python3 neuro_breakout.py
Will run a simple model I created using my own data, so it may not work well for you.
Myo Hello world
python3 myo_raw.py
Uses pygame to plot each of the 8 myo channels, useful for debugging.
You can gather training data by running python3 neuro_training.py.
Follow the paddle with your hand and the program will gather training data, for a default of 20 seconds before saving it to foo.csv.
Note that sEMG data, the same kind gathered by the Myo is thought to be uniquely identifiable. Do not share this data without careful consideration of the future implications.
python3 neuro_testing.py
Neuro_testing is a simple program made to test out different simple models.
Made as a weekend project, it is rough around the edges.
The sEMG sensor used was a Myo gesture control armband made by Thalmic Labs.
The Pygame breakout tutorial I used. Thanks 101 computing!
The code is primarily developed on Linux and should work on Windows and MacOS.
To communicate with the Myo, I used pyomyo.
emg_mode.PREPROCESSED (0x01)
By default myo-raw sends 50Hz data that has been rectified and filtered, using a hidden 0x01 mode.
emg_mode.FILTERED (0x02)
Alvipe added the ability to also get filtered non-rectified sEMG (thanks Alvipe).
emg_mode.RAW (0x03)
Then I further added the ability to get true raw non-filtered data at 200Hz. This data is unrectified but scales from -128 and 127.
Sample data and a comparison between data captured in these modes can be found in MyoEMGPreprocessing.ipynb. Also in this notebook is an explanation of the preprocessing done to the sEMG data and why we do it.
Apart from simpily the fun of it, many examples using the Myo are for discrete gesture recognition. Before moving onto a more complex regression task, I wanted an example of continuous gesture prediction / regression using sEMG data, this code serves as that example. It also shows how to use myo-raw in a seperate process and pass the data back using a queue.
In many statistical methods, the complexity of the model hinders it's ability to generalise.
With sEMG data there are two key generalisations:
Cross Session Generalisation: Many factors affect the data sent from the Myo. One key factor is the rotation of the Myo but other factors such as the temperature of the sensors affect the data.
Cross Person Generalisation:
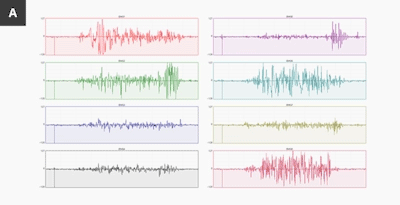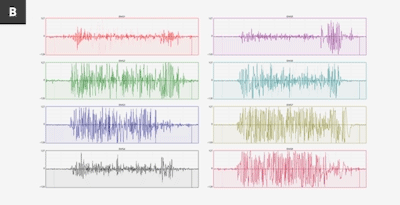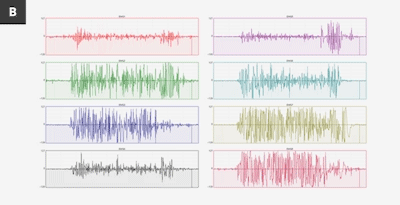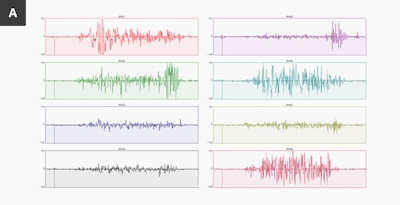
Above is an image from the Myo blog, showing data from two different people making the exact same gesture, any assumption we use in making a model may not apply to people outside of the same data.
This image hints at the uniquenes of sEMG data, like many things this could be used for good or bad. Possible uses include using sEMG as a password, or to fingerprint and track users, especially when combined with other data sources. As always, think before you share.
An obvious example is if the user is left or right handed. I realised another important factor when I tried a more complex model on someone with a much smaller wrist than mine, I assumed the Myo sizing clips would solve this problem, but they did not. From the collected data (ommitted for privacy) it seemed, the muscle I was targetting would span across 4 sensors on their wrist but only 2 on mine and therefore even with fixed rotation of the band I cannot assume which muscles show up in what channel. This gets more complicated when I considered the ratio between the fixed area of the sensor plate and the variable muscle size of different users.
Simplicity to the rescue!
This is why I use predict_simple which is a model that sums the amplitude of all the sensors, using rectified data that has been filtered for noise.
I made this model based on the size principle of motor unit recruitment, I expected to get higher amplitudes as the muscles were active as the right was pushed rather right.
This simple model is likely to need rescaling. Due to the filtering of the Myo, the sEMG data is no longer bounded from -128 - 127, therefore I manually scale to my own biology. Anyone else who tries this out will need to rescale to fit to the screen, I should use moving averages to allow the user to move the paddle the length of the screen and also rescale this band to deal with muscle fatigue as the user plays.