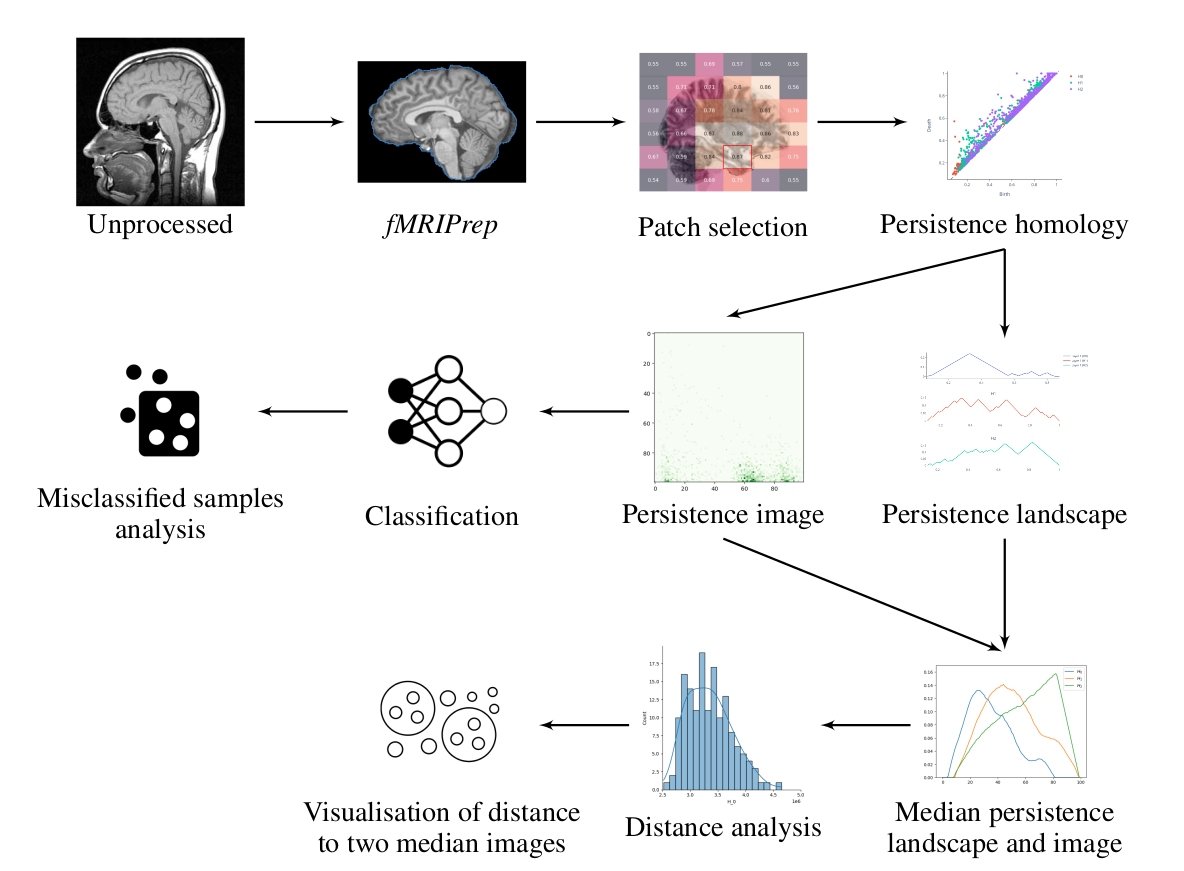
This is the repository containing a TDA of the ADNI data done in the context of a lab rotation at the MLCB lab of ETHZ. The goal of this project was fourfold:
- How salient are the topological features extracted from the patch for the characterization of the atrophy observed in Alzheimer’s disease as measured through the classification performance of AD versus CN subjects using persistence images?
- What does the distance between the persistence image and persistence landscape of a patient and the median persistence image or landscape of a diagnostic category reveal about the topological heterogeneity within a given diagnostic category?
- How can the distance among persistent images taken for each patient over the course of the disease inform us with regard to the progression of the patient during the monitoring period?
- Does taking the distance between each image and two median persistence images (from AD patients and CN subjects) allow adequate clustering of patients in diagnostic categories or disease subtypes?
All dependencies are managed through poetry. Use poetry install to install them.
This repository is organized as follows:
├── literature # Some useful literature
├── presentation # Presentation made to the lab on 2/12/2020
│ ├── figures
│ ├── lab_update.pdf
│ ├── lab_update.tex
│ └── References.bib
├── pyproject.toml # Poetry dependencies
├── README.org
├── report # Scripts in this directory answer question 4
│ ├── figures # Contains figures compiled for the report
│ ├── main.bib
│ ├── neurips_2020.sty
│ ├── report_lab_rotation.tex
└── scripts # Contains all scripts used in the analysis.
├── clustering # Scripts in this directory answer question 4
├── distances_from_median_rep # Scripts in this directory answer parts question 2
├── global # Yields data to train CNNs on whole-brain data
├── models # Answers Q1 (also contains models not discussed in the report)
├── outliers_misclassified # Answers Q2
├── patient_evolution # Answers Q3
├── persistent_homology # Applies persistent homology on the sMRI, useful for all questions.
├── plotting # Produces all the plots in figures/
└── utils.py # Utilities used by multiple scripts.
I would like to thank Bastian Rieck for the supervision, Catherine Jutzeler and Sarah Brüningk for helpful discussions, and Karsten Borgwardt for providing the research environment.