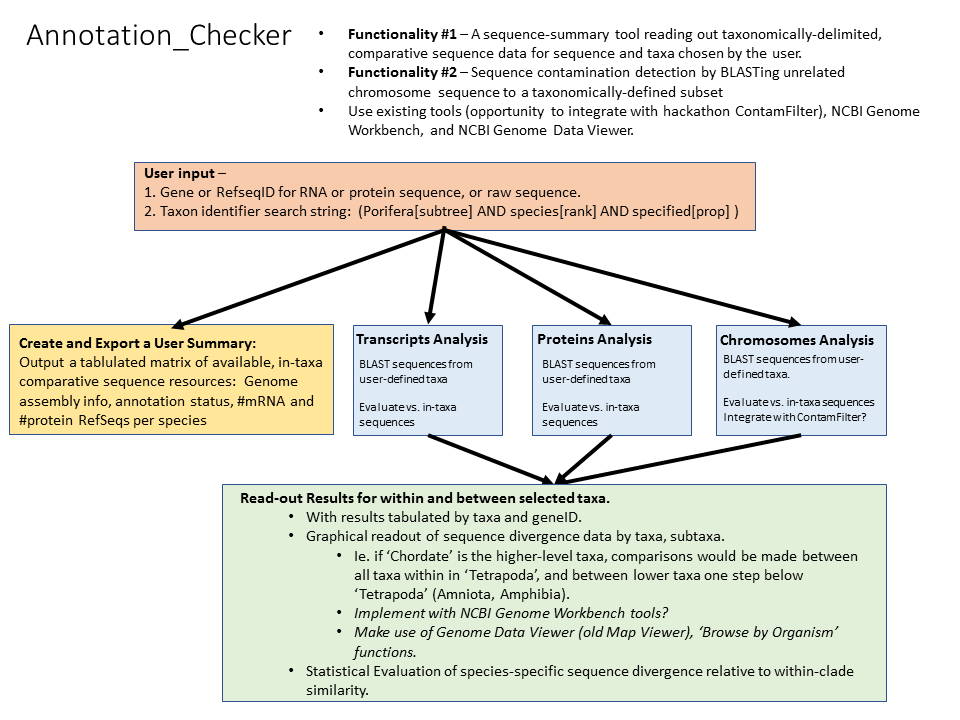
Annotation_Checker will provide a user-selectable, taxonomically subsetted, NCBI database-specific readout for identifying divergent taxonomic sequences. The tool will provide a quantitative capability to differentiate genome sequence quality from biologically-based evolutionary divergence.
11-17. Check each taxid for sequence database resources - 0/1? genomic DNA, RNA, protein databases, #accessions. Parse a subset of taxids possessing each resource.
$ esearch -db genome -query txid10088[Organism:exp] | esummary
$<!DOCTYPE DocumentSummarySet PUBLIC "-//NLM//DTD esummary genome 20140703//EN" "https://eutils.ncbi.nlm.nih.gov/eutils/dtd/20140703/esummary_genome.dtd">
<DocumentSummarySet status="OK">
<DbBuild>Build171117-1300.1</DbBuild>
<DocumentSummary><Id>53616</Id>
<Organism_Name>Mus pahari</Organism_Name>
<Organism_Kingdom>Eukaryota</Organism_Kingdom>
<Organism_Group></Organism_Group>
<Organism_Subgroup>Mammals</Organism_Subgroup>
<Defline></Defline>
<ProjectID>0</ProjectID>
<Project_Accession></Project_Accession>
<Status>Complete</Status>
<Number_of_Chromosomes>24</Number_of_Chromosomes>
<Number_of_Plasmids>0</Number_of_Plasmids>
<Number_of_Organelles>0</Number_of_Organelles>
<Assembly_Name>PAHARI_EIJ_v1.1</Assembly_Name>
<Assembly_Accession>GCA_900095145.2</Assembly_Accession>
<AssemblyID>1086041</AssemblyID>
<Create_Date>2017/01/10 00:00</Create_Date>
<Options></Options>
<Weight>0</Weight>
<Chromosome_assemblies>1</Chromosome_assemblies>
<Scaffold_assemblies>0</Scaffold_assemblies>
<SRA_genomes>0</SRA_genomes>
<TaxId>10093</TaxId>
</DocumentSummary>
<DocumentSummary><Id>34289</Id>
<Organism_Name>Mus caroli</Organism_Name>
<Organism_Kingdom>Eukaryota</Organism_Kingdom>
<Organism_Group></Organism_Group>
<Organism_Subgroup>Mammals</Organism_Subgroup>
<Defline>Mus caroli RefSeq Genome</Defline>
<ProjectID>264557</ProjectID>
<Project_Accession>PRJNA264557</Project_Accession>
<Status>Complete</Status>
<Number_of_Chromosomes>20</Number_of_Chromosomes>
<Number_of_Plasmids>0</Number_of_Plasmids>
<Number_of_Organelles>1</Number_of_Organelles>
<Assembly_Name>CAROLI_EIJ_v1.1</Assembly_Name>
<Assembly_Accession>GCA_900094665.2</Assembly_Accession>
<AssemblyID>1086031</AssemblyID>
<Create_Date>2017/01/10 00:00</Create_Date>
<Options></Options>
<Weight>0</Weight>
<Chromosome_assemblies>1</Chromosome_assemblies>
<Scaffold_assemblies>0</Scaffold_assemblies>
<SRA_genomes>0</SRA_genomes>
<TaxId>10089</TaxId>
</DocumentSummary>
<DocumentSummary><Id>12818</Id>
<Organism_Name>Mus spretus</Organism_Name>
<Organism_Kingdom>Eukaryota</Organism_Kingdom>
<Organism_Group></Organism_Group>
<Organism_Subgroup>Mammals</Organism_Subgroup>
<Defline>Mus spretus overview</Defline>
<ProjectID>83951</ProjectID>
<Project_Accession>PRJNA83951</Project_Accession>
<Status>Complete</Status>
<Number_of_Chromosomes>20</Number_of_Chromosomes>
<Number_of_Plasmids>0</Number_of_Plasmids>
<Number_of_Organelles>1</Number_of_Organelles>
<Assembly_Name>SPRET_EiJ_v1</Assembly_Name>
<Assembly_Accession>GCA_001624865.1</Assembly_Accession>
<AssemblyID>731341</AssemblyID>
<Create_Date>2016/04/15 00:00</Create_Date>
<Options></Options>
<Weight>0</Weight>
<Chromosome_assemblies>1</Chromosome_assemblies>
<Scaffold_assemblies>0</Scaffold_assemblies>
<SRA_genomes>0</SRA_genomes>
<TaxId>10096</TaxId>
</DocumentSummary>
<DocumentSummary><Id>52</Id>
<Organism_Name>Mus musculus</Organism_Name>
<Organism_Kingdom>Eukaryota</Organism_Kingdom>
<Organism_Group></Organism_Group>
<Organism_Subgroup>Mammals</Organism_Subgroup>
<Defline>The laboratory mouse is a major model organism for basic mammalian biology, human disease, and genome evolution, and its genome has been sequenced</Defline>
<ProjectID>9559</ProjectID>
<Project_Accession>PRJNA9559</Project_Accession>
<Status>Complete</Status>
<Number_of_Chromosomes>21</Number_of_Chromosomes>
<Number_of_Plasmids>0</Number_of_Plasmids>
<Number_of_Organelles>1</Number_of_Organelles>
<Assembly_Name>GRCm38.p6</Assembly_Name>
<Assembly_Accession>GCA_000001635.8</Assembly_Accession>
<AssemblyID>1198761</AssemblyID>
<Create_Date>2002/05/15 00:00</Create_Date>
<Options></Options>
<Weight>999</Weight>
<Chromosome_assemblies>17</Chromosome_assemblies>
<Scaffold_assemblies>5</Scaffold_assemblies>
<SRA_genomes>0</SRA_genomes>
<TaxId>10090</TaxId>
</DocumentSummary>
</DocumentSummarySet>
$ python3.6 Taxpull.py > taxids
##Taxpull.py
from ete3 import NCBITaxa
ncbi = NCBITaxa()
descendants = ncbi.get_descendant_taxa('Mus')
print(descendants)
$ [10089, 10091, 10092, 35531, 39442, 46456, 57486, 80274, 116058, 179238, 477815, 477816, 947985, 1266728, 1385377, 1643390, 1879032, 10096, 135827
, 135828, 10098, 270352, 270353, 10103, 27681, 83773, 186193, 186842, 254704, 473865, 481680, 10093, 41269, 10102, 10105, 41270, 60742, 273921, 27
3922, 397330, 397331, 397332, 397333, 544437, 887131, 1380868, 1582499, 1582501, 1582502, 1582503, 1582508, 1582509, 1890198, 10094, 10101, 210726
, 273920, 10095, 10108, 100383, 229288, 270354, 310402, 390847, 390848, 468371, 592722, 1390222, 1582479, 1582480, 1582481, 1582482, 1582483, 1582
504, 1582505, 1582506, 1582507]
20171115
-
Pull user-specified RefSeq genomes by NCBI Taxonomy Database taxID's (https://www.ncbi.nlm.nih.gov/taxonomy). ie. (Danio rerio).
-
Generate a list of taxID's from a given node (eg. taxID's for all formal species descending from 'Protostomia'.
-
Best tools available from Python-based ETE-Toolkit (http://etetoolkit.org/) , specifically the getting_descendant_taxa() command (http://etetoolkit.org/docs/2.3/tutorial/tutorial_ncbitaxonomy.html#getting-descendant-taxa), no quick alternatives appear elsewhere - may avoid "traversing the taxonomy database": (https://www.biostars.org/p/6528/) (https://www.biostars.org/p/16262/) (https://www.biostars.org/p/13452/)
Jaime Huerta-Cepas, Francois Serra and Peer Bork. ETE 3: Reconstruction, analysis and visualization of phylogenomic data. Mol Biol Evol 2016; doi: 10.1093/molbev/msw046
- Install ETE-toolkit. Windows-based Oracle virtual box or windows python?
20171114 -Began to work on user taxon and sequence selection.
-
Write a search string using NCBI EUtils to pull a reference sequence. NP_037367 = ribonuclease 3 isoform 1 [Homo sapiens]. https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=protein&term=NP_037367
-
Fetch the sequence fasta file. https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=protein&id=155030234&rettype=fasta&retmode=text
-
BLAST the reference fasta against user-selected taxID list. For testing and comparison, a list of 6 vertebrates (plus human reference) with well-validated genomic annotations, were chosen (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi). Each taxa was BLASTed against the user reference sequence, and the best hit was selected if BLAST result was valid:
taxIDs: 9913 - Bos taurus (domesticated cow) 7955 - Danio (zebrafish) 9606 - Human 10090 - Mouse 4932 - Rat 8355 - Takifugu (pufferfish) 4577 - Xenopus
- Continue working to automate this script.
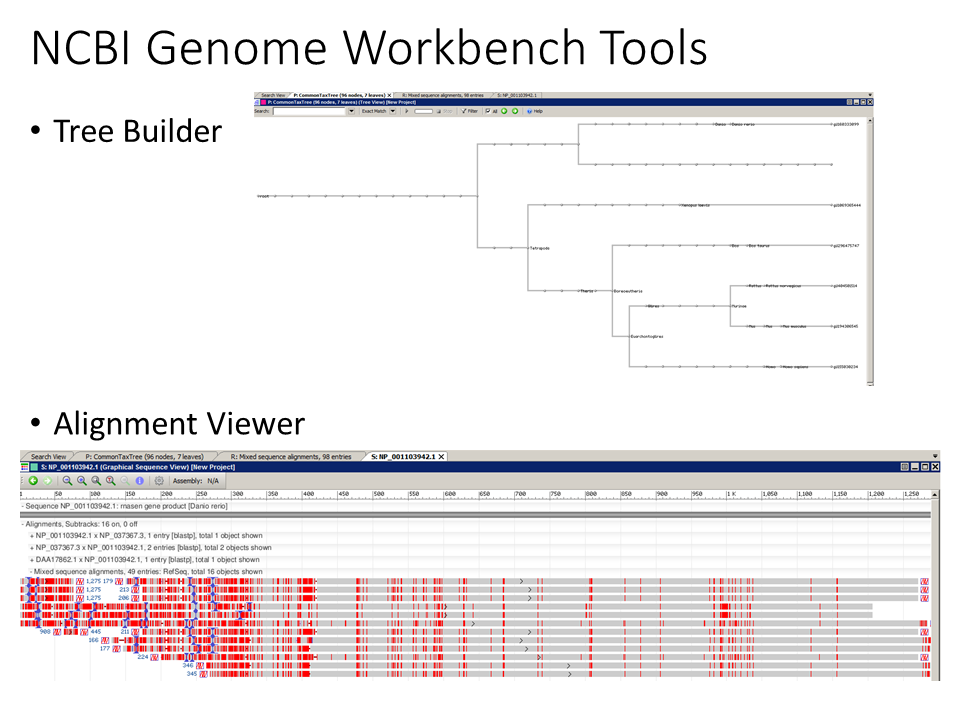
-NCBI Genome Workbench (https://www.ncbi.nlm.nih.gov/tools/gbench/) contains useful functionality. Work to incorporate useful features into Annotation_Checker User Summary readouts.