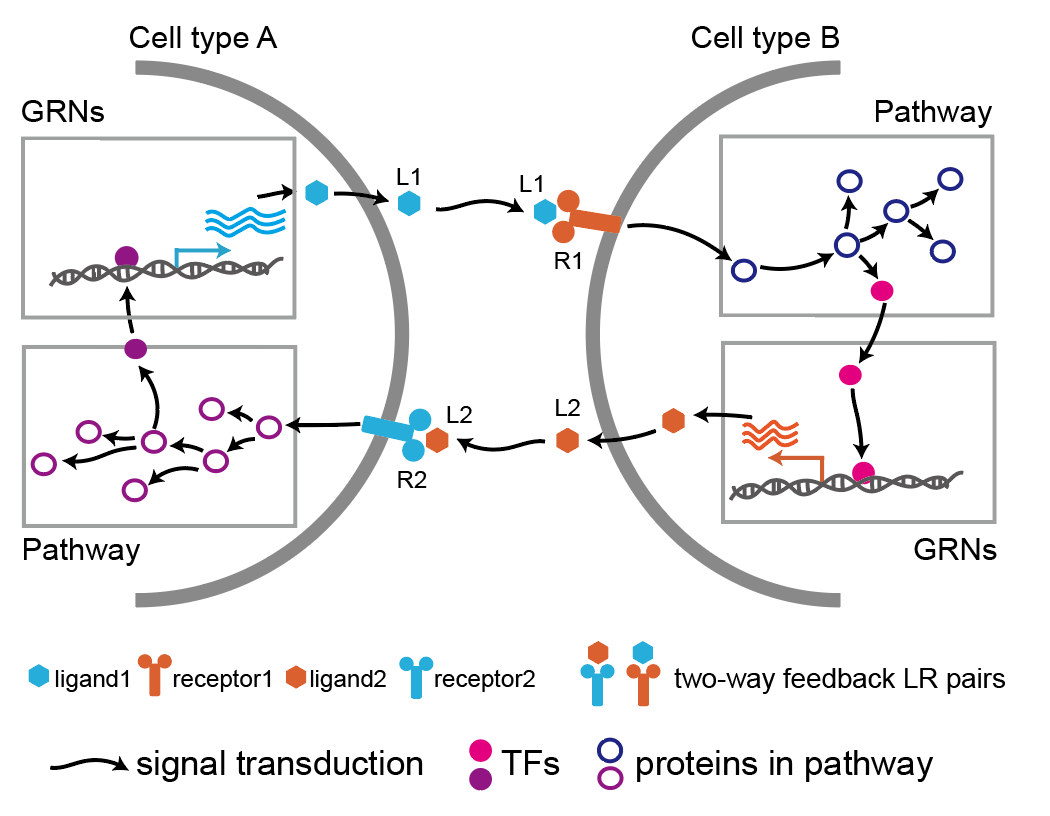
LRLoop is a full-featured R package for analyzing LR-Loops from bulk & single-cell RNA-seq data.
First, install devtools (for installing GitHub packages) if it isn't already installed
if (!requireNamespace("devtools", quietly = TRUE)) install.packages("devtools")
Then, install nichenetr if it isn't already installed
devtools::install_github("saeyslab/nichenetr")
Then, install all of the LRLoop dependencies
install.packages(c('tidyverse','Seurat','ggplot2','dplyr','circlize','igraph','RColorBrewer','writexl','pheatmap'))
Lastly, install LRLoop
devtools::install_github('https://github.com/Pinlyu3/LRLoop',force = TRUE)
To learn how to use LRLoop, read the following vignettes explaining several types of analyses:
-
The datasets in vignettes can be downloaded from google drive
LRLoop is currently in beta. If you think you have found a bug, please report an issue on Github with the Bug Report form.