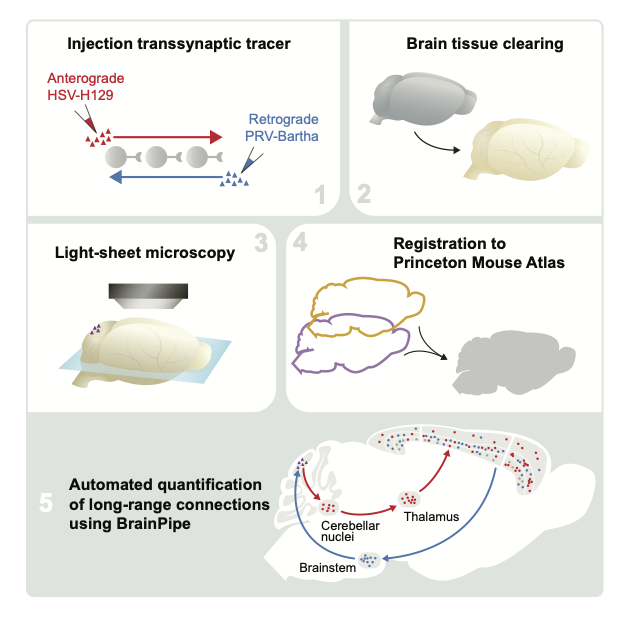
BrainPipe (Pisano et al. 2021) is an automated transsynaptic tracing pipeline that combines anatomical assignment using volumetric registration and injection site segmentation with convolutional neural networks for cell detection.
Volumetric alignment of brain volumes to the Princeton Mouse Atlas enables automated anatomical assignment
Please see INSTALLATION.md for installation instructions.
Please see EXAMPLES.md for basic BrainPipe use cases.
Demonstration datasets to ensure proper usage of BrainPipe and trained CNNs for H129 and PRV detection can be found here.
For CNN Demo: EXAMPLES: CNN Demo. Note, pre-trained CNN's (available here) are a good starting point for transfer learning, but probably will not work out-of-the-box for other projects' datasets. For a tutorial on making your own training set, see tutorials/make_UNet_training_set.ipynb.
For a deeper-dive into the BrainPipe pipeline, please see IMPORTANT_FILES.md for details. When starting to use your own data, BrainPipe expects certain formatting of images (see FILE-FORMATTING.md for details).
Convolutional neural networks efficiently and accurately detect virally-labeled neurons in whole cleared mouse brains
Princeton Mouse Atlas: a volumetric atlas with a complete cerebellum that is compatible with the Allen Brain Atlas
The Princeton Mouse Atlas (PMA) was generated from 110 light-sheet volumes and is compatible with the Allen Brain Atlas. PMA data can be found here. Further PMA details can be found in Pisano et al. 2021.
 Aligned anterograde (HSV-H129) and retrograde (PRV-Bartha) viral tracing injection data from Pisano et al. 2021 has been deposited here.
Aligned anterograde (HSV-H129) and retrograde (PRV-Bartha) viral tracing injection data from Pisano et al. 2021 has been deposited here.