This repository is the official implementation of our NeurIPS 2023 paper "Unsupervised Polychromatic Neural Representation for CT Metal Artifact Reduction" [OpenReview], [arXiv]
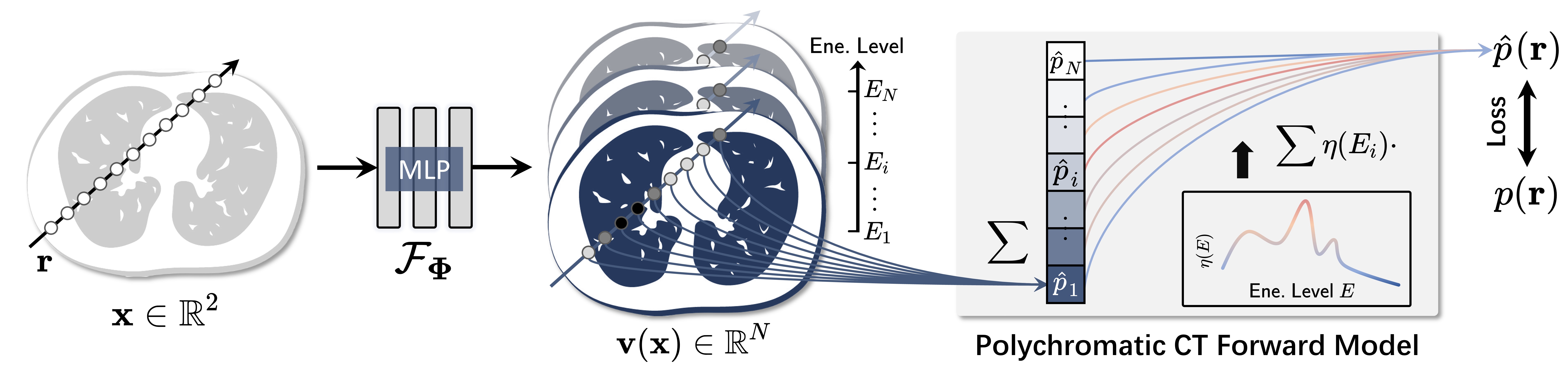 Fig. 1: Overview of the proposed Polyner model.
Fig. 1: Overview of the proposed Polyner model.
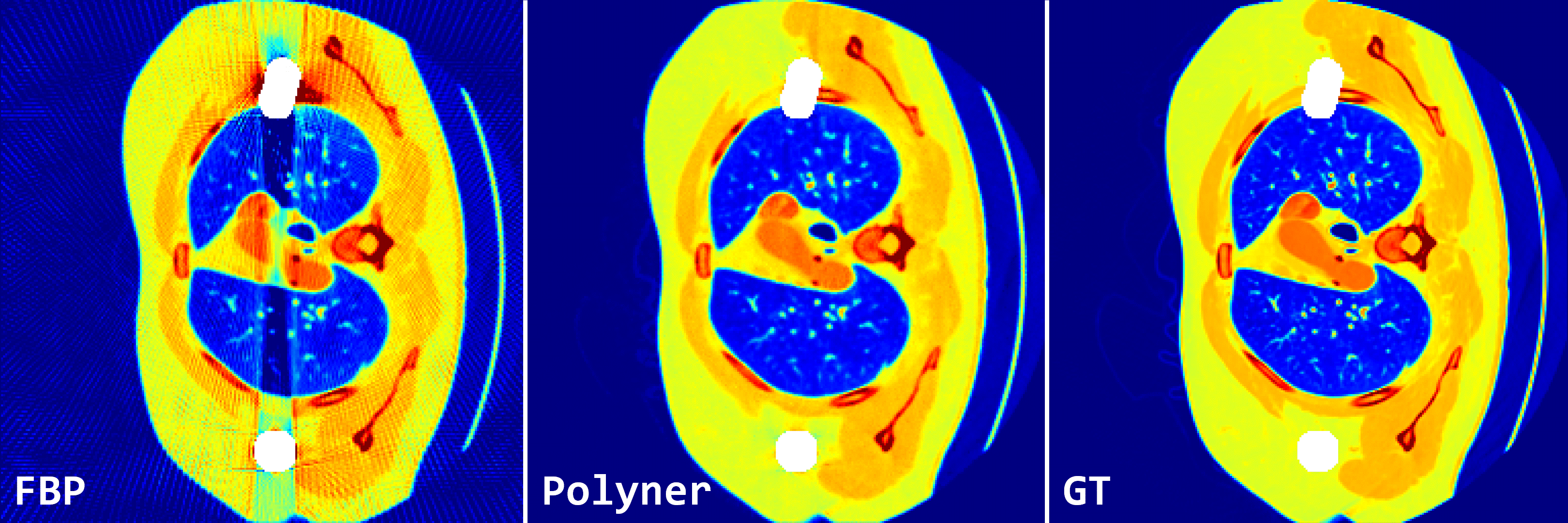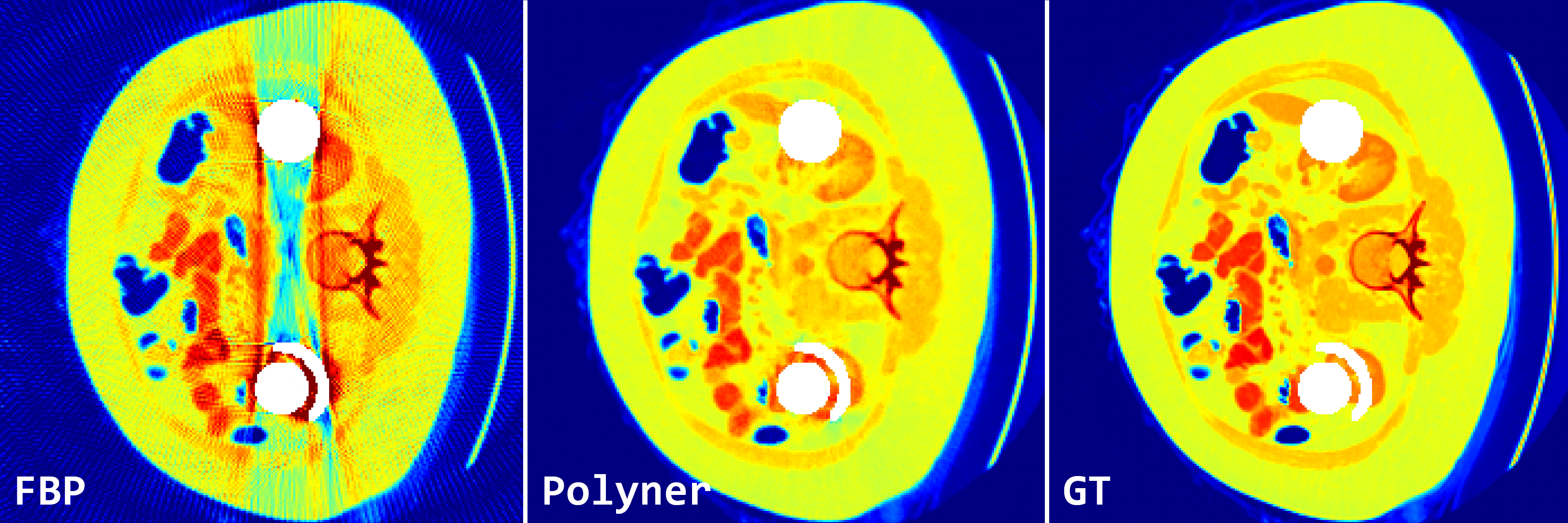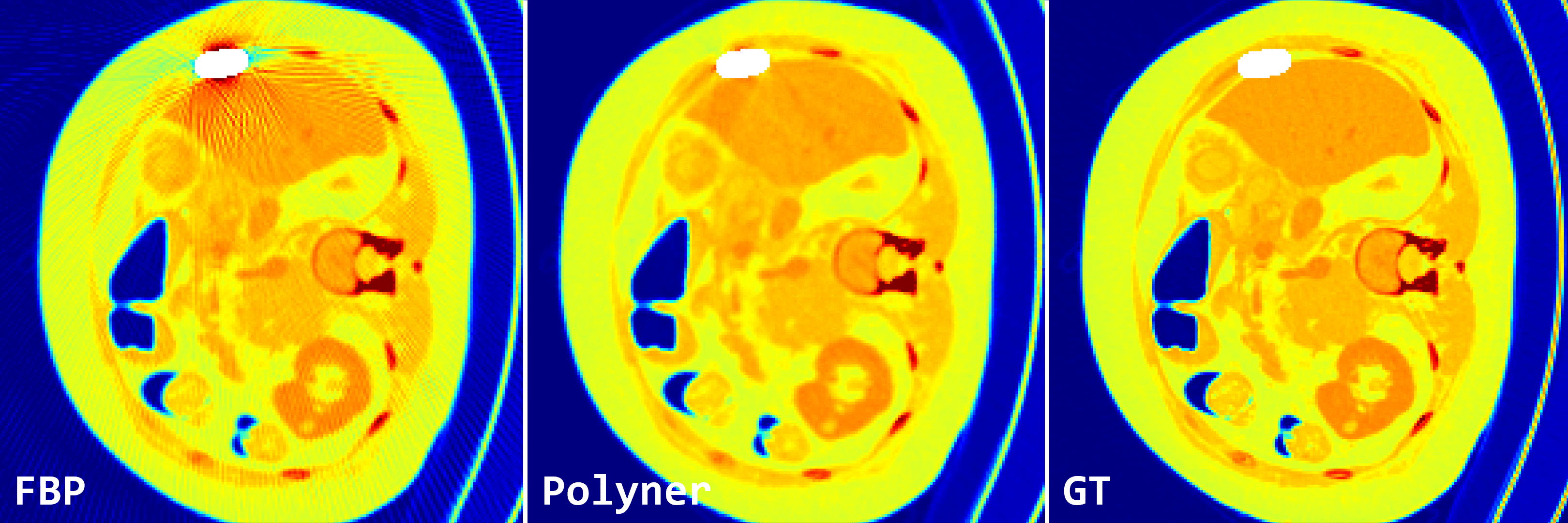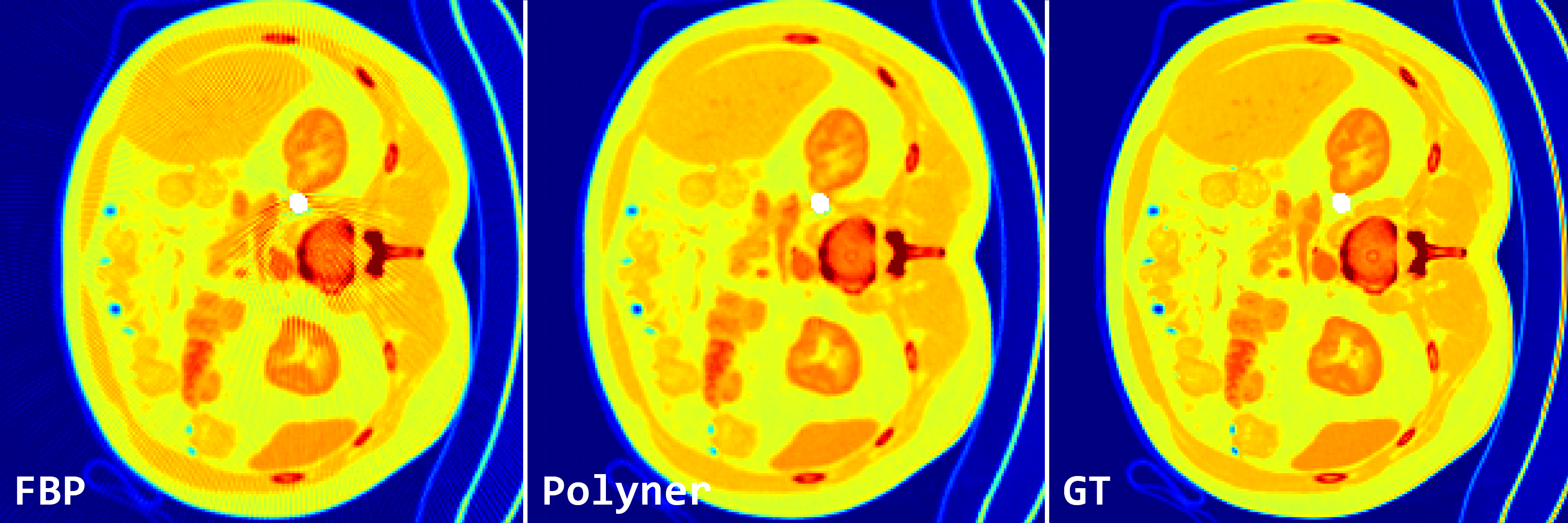 Fig. 2: Qualitative results of FBP and our polyner on 2D fan-beam samples of DeepLesion simulation dataset.
Fig. 2: Qualitative results of FBP and our polyner on 2D fan-beam samples of DeepLesion simulation dataset.
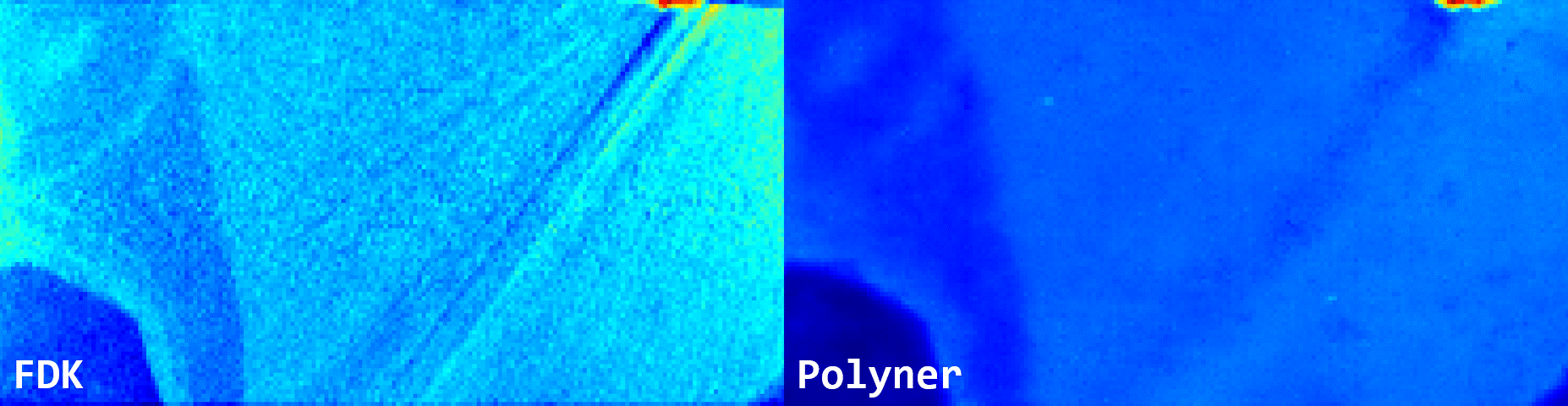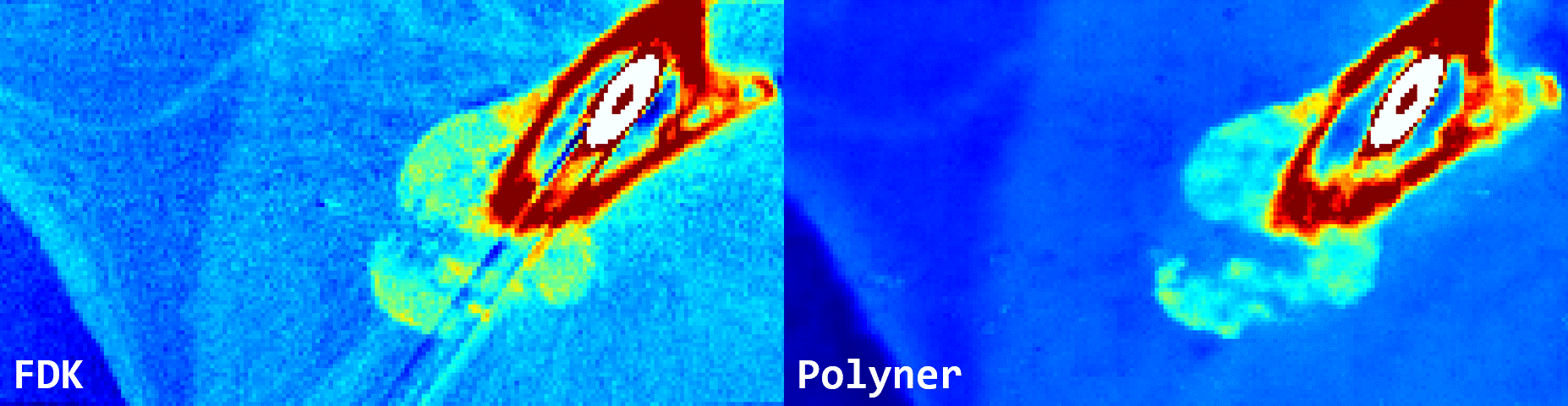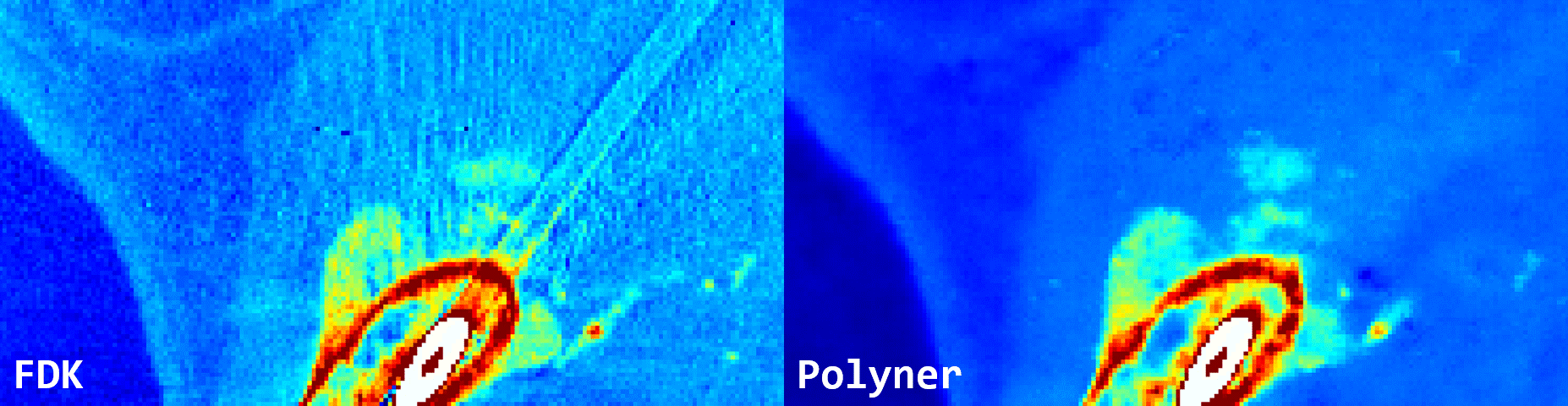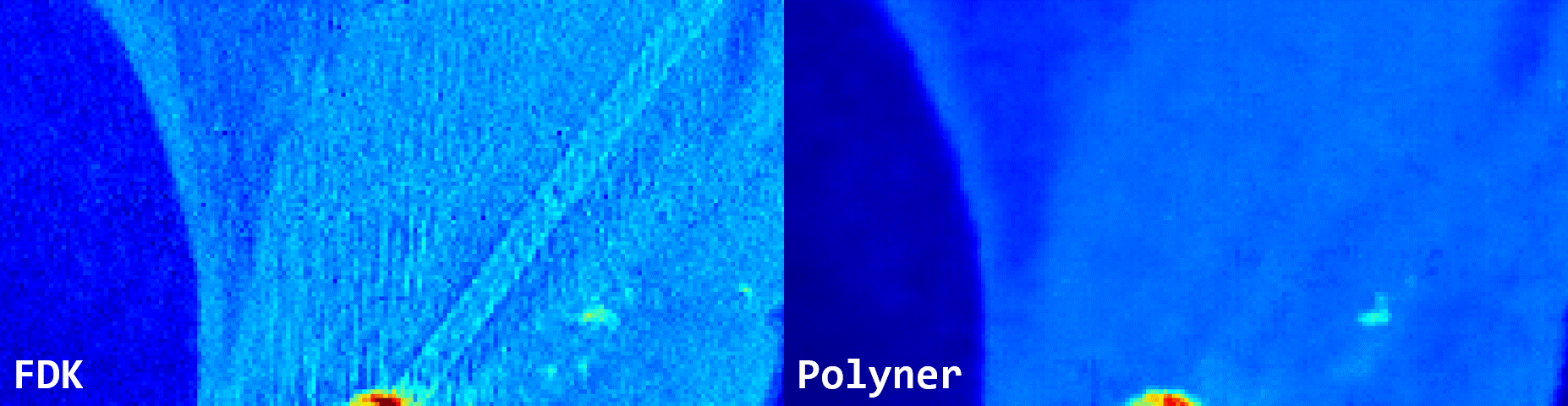 Fig. 3: Qualitative results of FDK and our polyner on a real-world 3D cone-beam mouse thigh sample.
Fig. 3: Qualitative results of FDK and our polyner on a real-world 3D cone-beam mouse thigh sample.
Polyner
│ config.json # configuration script.
│ dataset.py # dataloader
│ eval.py # quantitative evaluation
│ main.py # running script for training
│ model.py # EAS loss
│ readme.md # readme file
│ Polyner.py # training function
│ utils.py # tools
│
├─data_simulation # data simulation
│ │ config_dl.yaml # acquisition parameters
│ │ dl_data.m # running script for DeepLesion dataset
│ │
│ ├─+helper # functions for data simulation
│ │ get_mar_params.m
│ │ interpolate_projection.m
│ │ pkev2kvp.m
│ │ simulate_metal_artifact.m
│ │
│ ├─metal # prior data for simulation
│ │
│ └─slice
│ gt_x.nii # raw data
│
├─input
│ fanSensorPos.nii # geometry angle
│ GE14Spectrum120KVP.mat # energy spectrum
│ gt_x.nii # gt image
│ mask_x.nii # metal mask
│ ma_x.nii # FBP reconstructions
│ ma_sinogram_x.nii # metal-corrupted measurements
│
├─model
│ model_x.pkl # pre-trained Polyner
│
└─output
polyner_x.nii # Polyner reconstructions
To run this project, you will need the following packages:
- PyTorch 3.8.13
- tinycudann
- SimpleITK, tqdm, numpy, and other packages.
To train our Polyner from scratch, navigate to ./ and run the following command in your terminal:
python main.pyThis will train the Polyner model for the metal-corrputed sinogram (./input/ma_sinogram_0~9.nii). The well-trained model will be stored in ./model and its corresponding MAR results will be stored in ./output.
To qualitatively evalute the result, navigate to ./ and run the following comman in your terminal:
python eval.pyThis will compute PSNR and SSIM values of FBP and our Polyner on the ten samples of the DeepLesion dataset.
For the ten sinograms (./input/ma_sinogram_0~9.nii), the quantitative results are shown in:
| Method | PSNR | SSIM |
|---|---|---|
| FBP | 29.13±3.27 | 0.7201±0.1109 |
| Polyner | 37.33±0.93 | 0.9774±0.0031 |
To simulate the metal-corrupted measurements, navigate to ./data_simulation and run the MATLAB script dl_data.m. These code for data simulation are based on the ADN repository: https://github.com/liaohaofu/adn/tree/master
NIFTI files (.nii) can be viewed by using the ITK-SNAP software, which is available for free download at: http://www.itksnap.org/pmwiki/pmwiki.php?n=Downloads.SNAP4
This code is available for non-commercial research and education purposes only. It is not allowed to be reproduced, exchanged, sold, or used for profit.
If you find our work useful in your research, please cite:
@inproceedings{
wu2023unsupervised,
title={Unsupervised Polychromatic Neural Representation for {CT} Metal Artifact Reduction},
author={Qing Wu and Lixuan Chen and Ce Wang and Hongjiang Wei and S Kevin Zhou and Jingyi Yu and Yuyao Zhang},
booktitle={Thirty-seventh Conference on Neural Information Processing Systems},
year={2023},
url={https://openreview.net/forum?id=xx3QgKyghS}
}