This R library facilitates the analysis of the difference between the proprotion of cells in clusters between two scRNA-seq samples. A permutation test is used to calculate a p-value for each cluster, and a confidence interval for the magnitude difference is returned via bootstrapping. There is also a function to generate a point range plot to display the results.
Use devtools to install the R library.
devtools::install_github("rpolicastro/scProportionTest")
This library pulls the meta-data from a seurat object for its analysis. This means that you must first process your data in seurat. Seurat has various vignettes to get you started.
Once you have a seurat object, you are ready to get started. We'll first load some example data for the vignette, and create the analysis object.
library("scProportionTest")
seurat_data <- system.file("extdata", "example_data.RDS", package = "scProportionTest")
seurat_data <- readRDS(seurat_data)
prop_test <- sc_utils(seurat_data)
Once the object is created, the permutation testing and bootstrapping can be run.
prop_test <- permutation_test(
prop_test, cluster_identity = "custom_clusters",
sample_1 = "HT29_EV", sample_2 = "HT29_LSD1_KD",
sample_identity = "orig.ident"
)
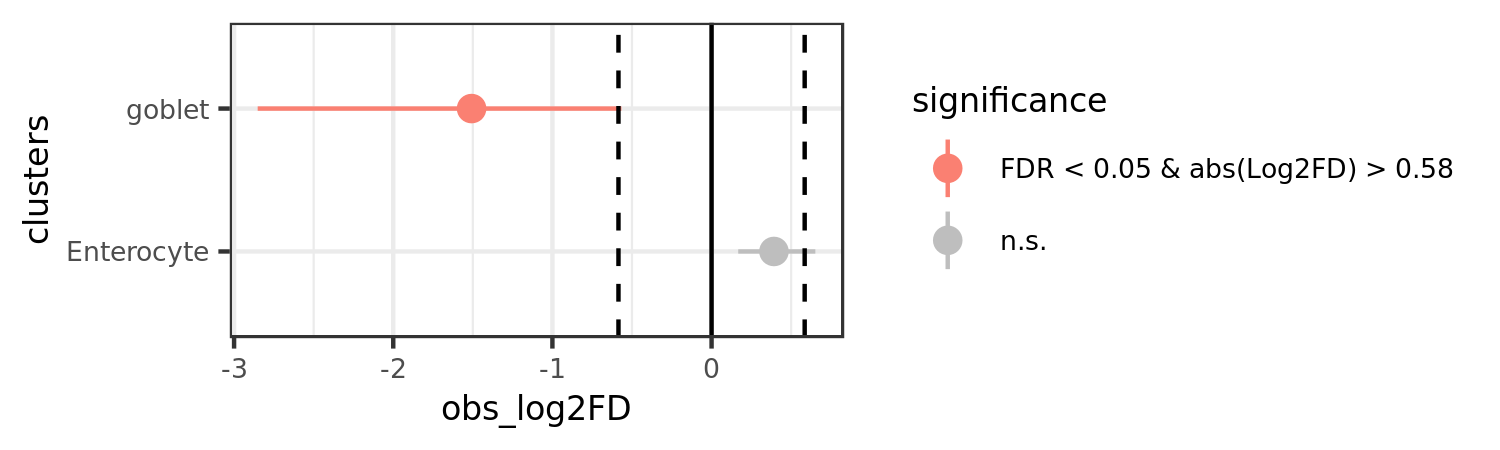
A point-range plot of the results can then be created.
permutation_plot(prop_test)