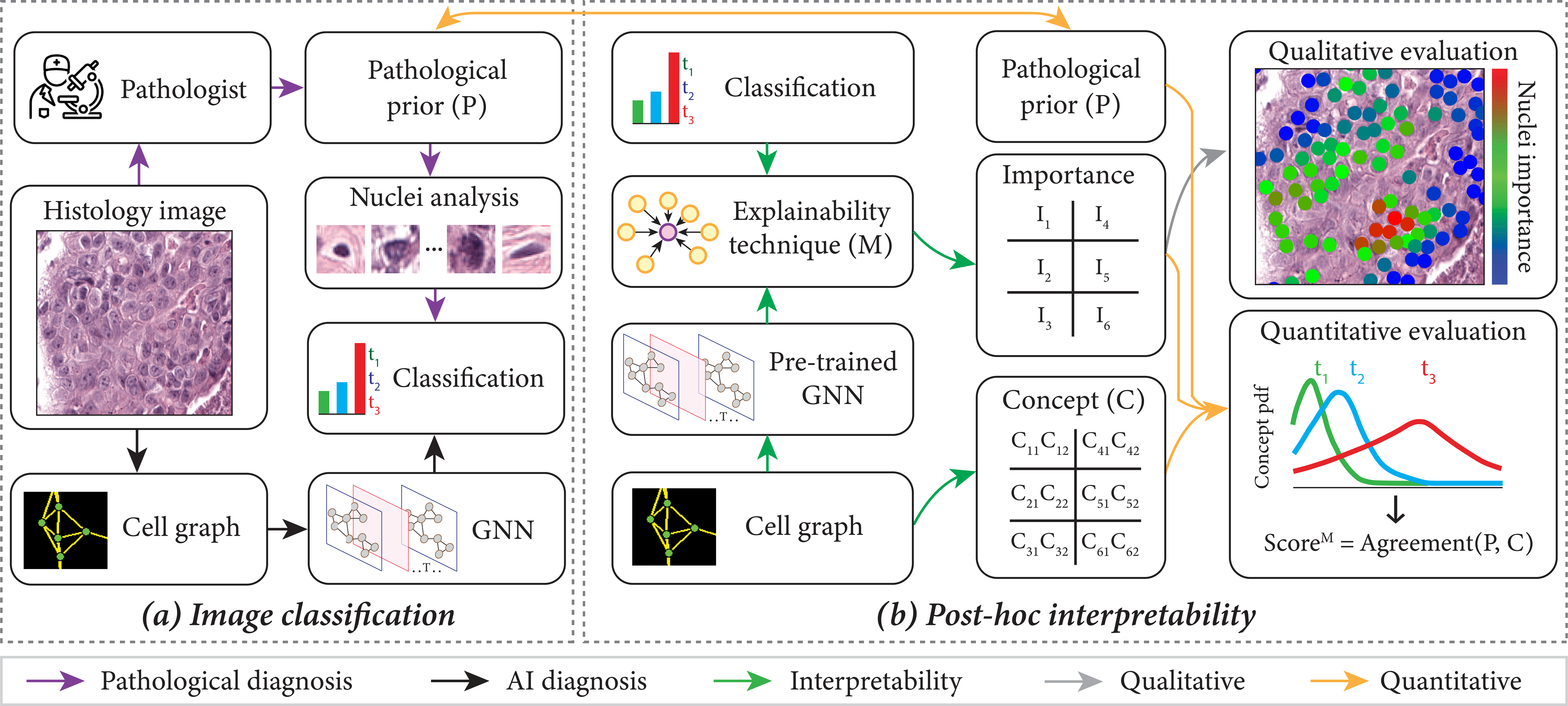
This repository includes the code for replicating results presented in the paper Quantifying Explainers of Graph Neural Networks in Computational Pathology presented at CVPR 2021.
The code mostly relies on the histocartography library, a python-based package for modeling and learning with graph-based representations of pathology images.
All the experiments are based on the BRACS dataset. The data needs to be downloaded separately (see Installation steps).
Clone the repo:
git clone https://github.com/histocartography/patho-quant-explainer.git && cd patho-quant-explainer
Create a conda environment and activate it:
conda env create -f environment.yml
conda activate pathoexplainer
Make sure that you have the latest version of histocartography, histocartography==0.2.
BRACS is a dataset of Hematoxylin and Eosin (H&E) histopathological images for automated detection/classification of breast tumors. BRACS includes >4k tumor regions-of-interest labeled in 7 categories (Normal, Benign, UDH, ADH, FEA, DCIS, Invasive).
In order to download the BRACS dataset, you need to create an account there. Then, go to Data Collection, Download, and hit the Regions of Interest Set button to access the data. Download the v1 data. The data are stored on an FTP server.
The approach for explainability of histology images is based on 3 steps: cell graph generation, post-hoc graph explaination to identify salient nodes and the proposed quantitative analysis.
Each image needs to be transformed into a cell graph where nodes represent nuclei and edges nuclei-nuclei interactions. The cell graph for the BRACS test set can be generated with:
cd core
python generate_cell_graphs.py --data_path <PATH-TO-BRACS>/BRACS/test/ --save_path <SOME-SAVE-PATH>/quant-gnn-explainers-data
The script will automatically create a directory containing cell graphs as a .bin file for each image. There should be 626 files created.
We benchmark 4 different explainers: GraphLRP, GNNExplainer, GraphGradCAM and GraphGradCAM++, that returns a different explanation, i.e., nuclei-level importance scores, for each cell graph. The system will automatically download a pre-trained checkpoint trained on 3-class BRACS using the histocartography library. This model reaches 74% accuracy on the test set.
Generating explanation with:
python generate_explanations --cell_graphs <SOME-SAVE-PATH>/quant-gnn-explainers-data/cell_graphs --explainer graphgradcam --save_path <SOME-SAVE-PATH>/quant-gnn-explainers-data/graphgradcam
The explainer type can be set to either: graphgradcam, graphgradcampp, gnnexplainer or graphlrp.
We provide a script to visualize the explanation by overlaying the node-level importance scores on the original image. For instance by running:
python visualize_explanations --cell_graphs <SOME-SAVE-PATH>/quant-gnn-explainers-data/cell_graphs --images <SOME-SAVE-PATH>/quant-gnn-explainers-data/images --importance_scores <SOME-SAVE-PATH>/quant-gnn-explainers-data/graphgradcam
--save_path <SOME-SAVE-PATH>/quant-gnn-explainers-data/visualization
If you don't wanna re-generate all the cell graphs and explanations for the whole set, we provide a zip file that you can directly download here. This file contains pre-processed cell graphs and explanations.
To run the quantitative analysis, you can simply run:
python run_qualitative_analysis.py --cell_graphs <SOME-SAVE-PATH>/quant-gnn-explainers-data/cell_graphs --importance_scores <SOME-SAVE-PATH>quant-gnn-explainers-data/graphgradcam --save_path <SOME-SAVE-PATH>/visualization/graphgradcam
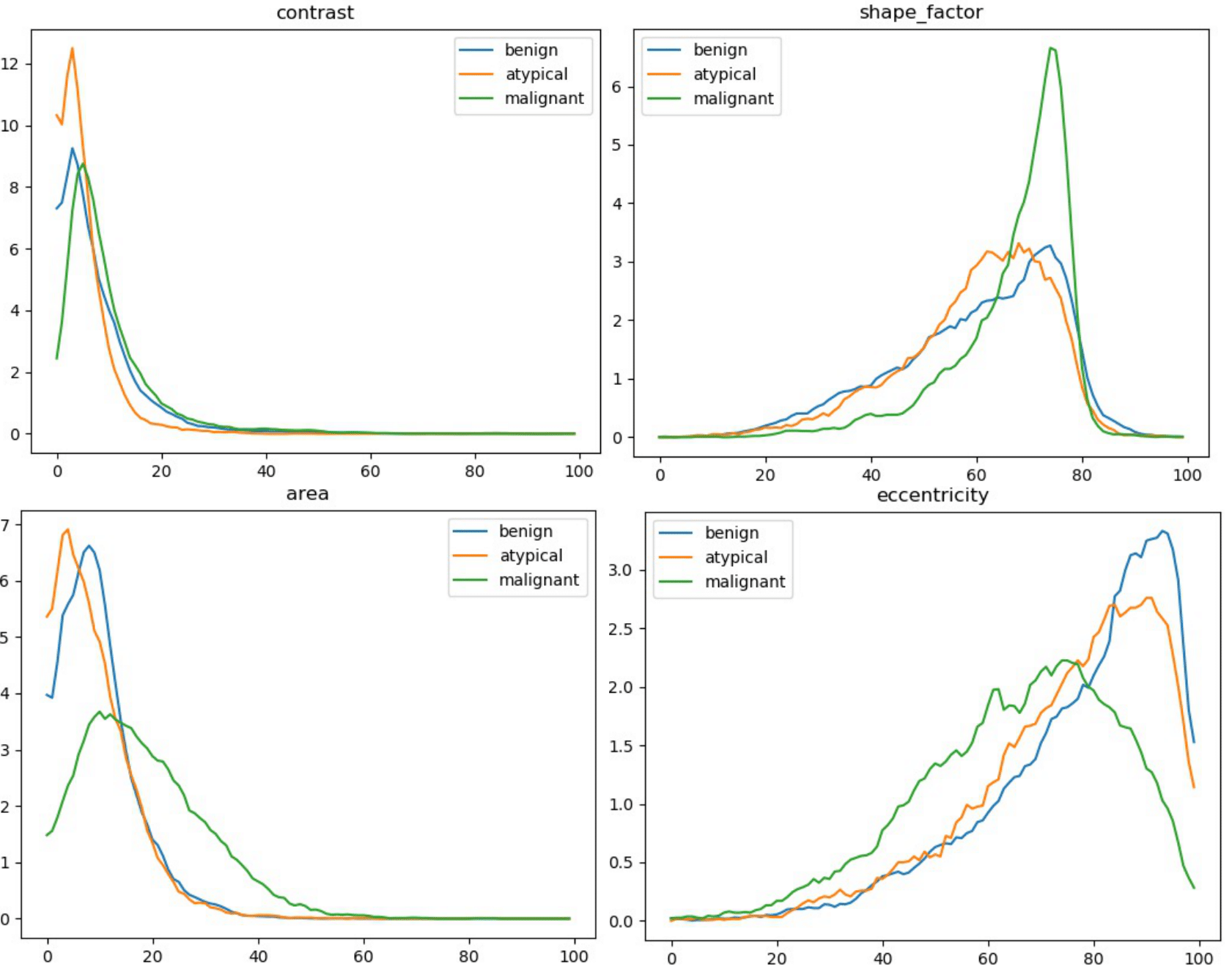
The code will save a number of histograms that represent the distribution of nuclei-level attributes per tumor type. The code also print the separability scores: average, maximum and correlation.
Note: The code will by default remove misclassified samples, it is also possible to run on the entire test set by adding the flag misclassification (see implementation for details).
An example of attribute histograms would be:
A number of elements changed in the pipeline explaining different numbers than the ones reported in the original paper. However, the conclusion remains. The differences include a different nuclei detector, an optimized stain normalization, a different list of nuclei-level attributes (changed to reduce computational requirements), a more robust attribute normalization.
If you use this code, please consider citing our work:
@inproceedings{jaume2021,
title = "Quantifying Explainers of Graph Neural Networks in Computational Pathology",
author = "Guillaume Jaume, Pushpak Pati, Behzad Bozorgtabar, Antonio Foncubierta-Rodríguez, Florinda Feroce, Anna Maria Anniciello, Tilman Rau, Jean-Philippe Thiran, Maria Gabrani, Orcun Goksel",
booktitle = "IEEE CVPR",
url = "https://arxiv.org/abs/2011.12646",
year = "2021"
}