SynAnno is a tool designed for proofreading and correcting synaptic polarity annotation from electron microscopy (EM) volumes - specifically the H01 dataset. SynAnno is aimed for integration with CAVE (Connectome Annotation Versioning Engine).
Harvard's Lichtman laboratory and Google's Connectomics team released the H01 dataset, a 1.4 petabyte view of human brain tissue via nanoscale EM. It covers a volume of ~1mm³, featuring tens of thousands of neurons, millions of neuron fragments, 183 million annotated synapses, and 100 proofread cells.
Synaptic polarity refers to the directionality of information flow between two neurons at a synapse, the junction where they communicate. In this context, we classify synapses as pre-synaptic (information senders) or post-synaptic (information receivers). A pre-synaptic neuron sends neurotransmitter signals across the synaptic cleft to the post-synaptic neuron, which receives these signals and processes the information. While the segmentation masks highlight the synaptic clefts between neurons, the pre-/post-synaptic IDs are coordinates placed into the associated neurons, identifying the specific sender and receiver. Identifying these key elements is crucial for creating accurate structural and functional neural maps. SynAnno assists in proofreading, correcting, and identifying these segmentation masks and synaptic IDs.
CAVE is a computational infrastructure to host petabyte connectomes for distributed proofreading and dynamic spatial annotation. Soon, we will provide support to use SynAnno together with CAVE-hosted datasets.
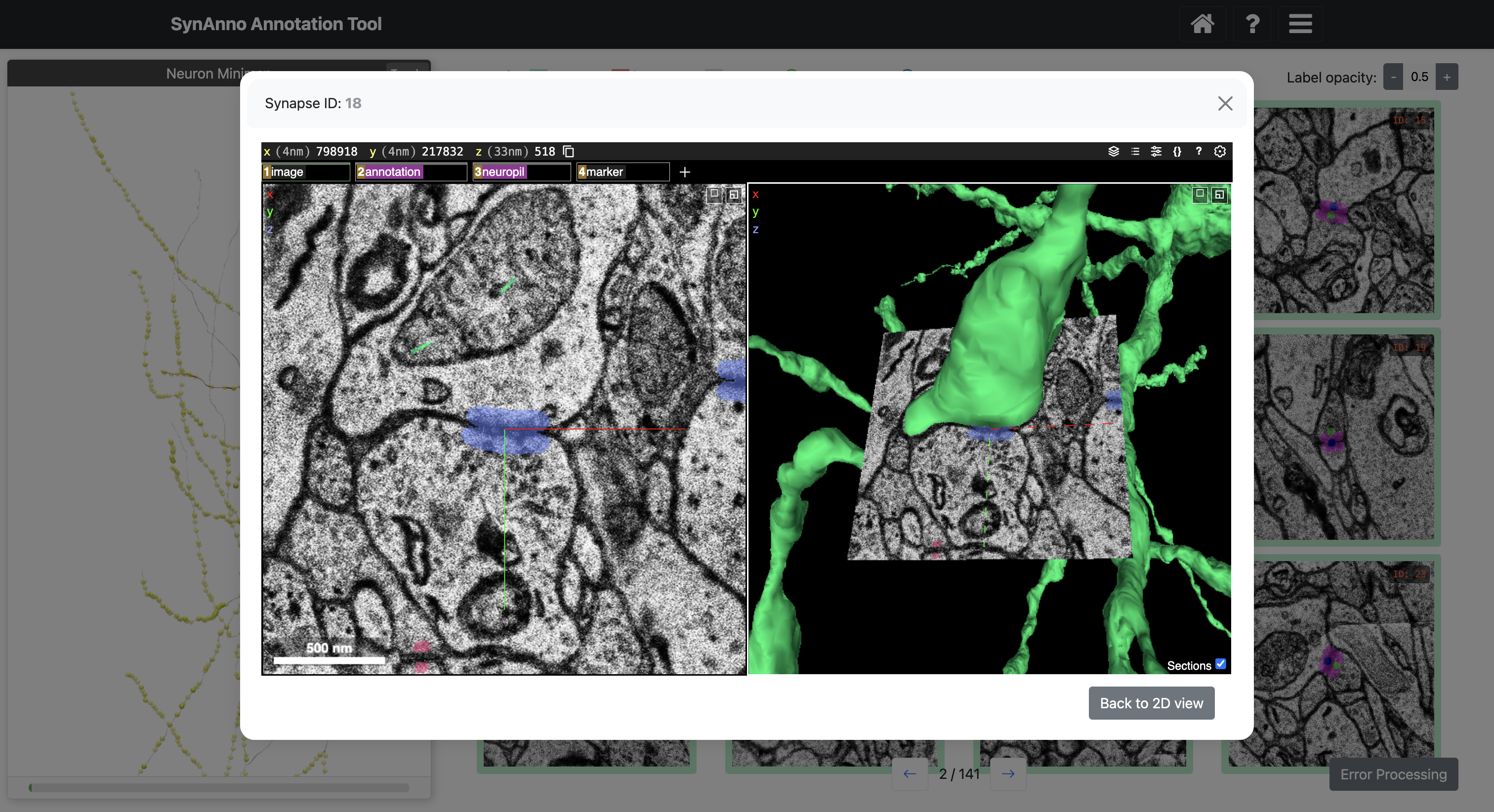
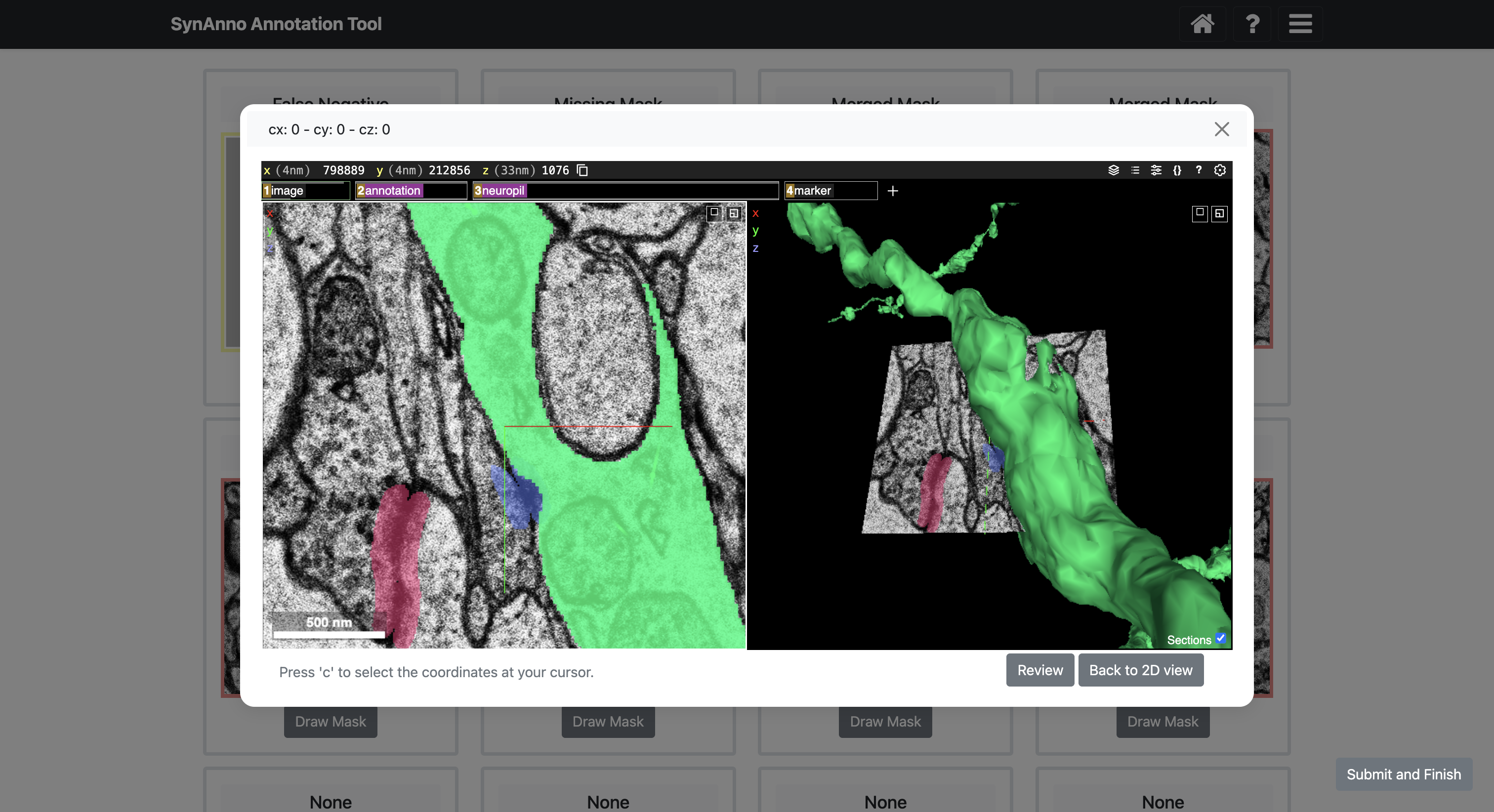
SynAnno integrates Neuroglancer directly into its interface. Neuroglancer is a powerful tool for 3D visualization of large-scale neuroimaging data. This integration allows users to effortlessly transition to a 3D view from any instance in the proofreading or drawing views. When an instance is selected, the embedded Neuroglancer opens at the exact location, providing an enhanced view of the specific instance. This functionality is particularly helpful during proofreading, enabling users to closely examine complex cases and make more informed decisions. In the drawing view, users can navigate through the dataset with ease, mark false negatives with a single click, and then return to SynAnno to draw segmentation masks, set pre-/post-synaptic coordinate IDs, or more accurately assess and correct erroneous cases when editing existing masks and IDs.
Leveraging CloudVolume, SynAnno efficiently handles vast datasets, such as the H01 1.4 petabyte volume, by employing on-demand, page-wise loading of instance-specific subvolumes. Users can seamlessly access synapses associated with specific pre and/or post-synaptic IDs or within designated subvolumes, allowing for the referencing of an unlimited number of neurons. SynAnno only retains metadata for each page and image data for instances marked as erroneous, optimizing memory usage. This targeted data retention enables quick reloading of problematic instances for further analysis and correction in the categorization and drawing views.
SynAnno's mask layout adheres to the H01 dataset standards, using a monochrome segmentation mask to highlight the synaptic cleft. In the proofreading view, pre-synaptic coordinate IDs are marked by a green dot, and post-synaptic coordinate IDs by a blue dot, with these markers presented in bright colors on their specific slice and in muted shades on related slices for easy reference. In the drawing mode, users have the flexibility to place pre-/post-synaptic ID markers on any slices independently, making it possible to accommodate synapses with varying orientations in the Neuroglancer view. The user can redraw mismatches by setting an adjustable number of spline points. Users can download the corrected segmentation mask directly, while pre-/post-synaptic IDs are stored in a pandas DataFrame, available for download as a JSON file.
The Materialization Table functions as a database that links annotations to segmentation IDs within large-scale neuroimaging datasets. It regularly updates based on the bound spatial points of the annotations and the underlying segment IDs, creating a systematic connection between annotations and IDs. This enables efficient querying for specific annotations or IDs, providing essential information for tracking connectivity in the datasets.
In SynAnno, the Materialization Table is simply a reference for determining which instances to load. SynAnno queries the table with the provided pre-/post-synaptic IDs or subvolume coordinates and loads the relevant instances based on the retrieved information. This approach streamlines the loading process, making it efficient and straightforward to access the required data.
-
Proofreading Annotated Data:
- View individual data instances, their associated segmentation masks and pre-/post-synaptic coordinate IDs.
- Mark segmentation masks and pre-/post-synaptic coordinate IDs that appear to be erroneous and Provide errors descriptions.
-
Segmentation Mask Corrections:
- Delete false positives.
- Add missed FN by browsing and marking them via Neuroglancer.
- Redraw mismatches using spline interpolation with intuitive control points.
- Reset pre-/post-synaptic coordinate IDs.
-
Cloud and Dataset Compatibility:
- Full integration of Neuroglancer's "precomputed" dateformat.
- Neuron-centric data loading: Instance based loading via pre-/post-synaptic IDs.
- View-centric data loading: Load all instance with in a given sub-volume range.
- Handle shapes and resolutions mismatches between source and target volumes.
- Support for arbitrary coordinate system (e.g., xyz, zyx).
-
Advanced Instance Management:
- 2D slice-wise navigation through all instances source and target slices.
- Instantly view instances in Neuroglancer for 3D analysis.
-
Efficient Data Handling:
- On-demand loading of instances using CloudVolume, suitable for large datasets.
- Reduce loading time for multiple instances through multi-threading.
- Centralize data management with a unified Pandas dataframe.
-
Future Features
- Depth-wise auto-segmentation via custom seed segmentation masks (see issue #70).
Download SynAnno and unzip it into a folder of your choice. For the following we assume you've unzipped the folder under /home/user/SynAnno. You can either run SynAnno in a Docker container or set up a local environment.
Repository includes a Dockerfile that enables you to build and run the application in a Docker container to isolate it from your local setup. It ensures a consistent environment across different machines, simplifying deployment and avoiding potential configuration issues.
-
Install Docker: If you haven't already, install Docker on your machine.
-
Navigate to the project folder: Open a terminal and navigate to the folder containing the Dockerfile.
cd /home/user/SynAnno -
Build the Docker image: Build the Docker image using the provided Dockerfile.
docker build -t synanno . -
Run the Docker container: Run the Docker container using the image you just built.
docker run -p 8080:80 synanno
This command maps port 8080 on your local machine to port 80 on the Docker container.
-
Access the application: Open a web browser and go to
http://localhost:8080to access the application running in the Docker container. -
Stop the Docker container: When you're done, you can stop the Docker container by pressing
Ctrl + Cin the terminal where the container is running.
Set up the environment of your choice and install the required packages from either the requirements.txt or Pipfile. In the following, we quickly outline how to derive a strong setup using pyenv and pipenv on macOS.
Why Use pyenv and pipenv?
- Isolation: Prevent dependency conflicts with isolated virtual environments.
- Reproducibility: Ensure consistent dependency versions with
Pipfile.lock. - Python Version Management: Easily switch Python versions per project.
-
Install
pyenv:brew install pyenv
-
Add
pyenvto your shell:echo 'eval "$(pyenv init --path)"' >> ~/.zshrc
-
Install Python version with
pyenv:pyenv install 3.9.12
-
Set global Python version:
pyenv global 3.9.12
-
Install
pipenv:pip install pipenv
-
Navigate to your project folder:
cd /home/user/SynAnno -
Initialize
pipenvenvironment:pipenv --python $(pyenv which python) -
Activate the environment:
pipenv shell
-
Install the
Pipfile:pipenv install
From with in the repository (e.g. /home/user/SynAnno) start SynAnno using the following command:
python run.py
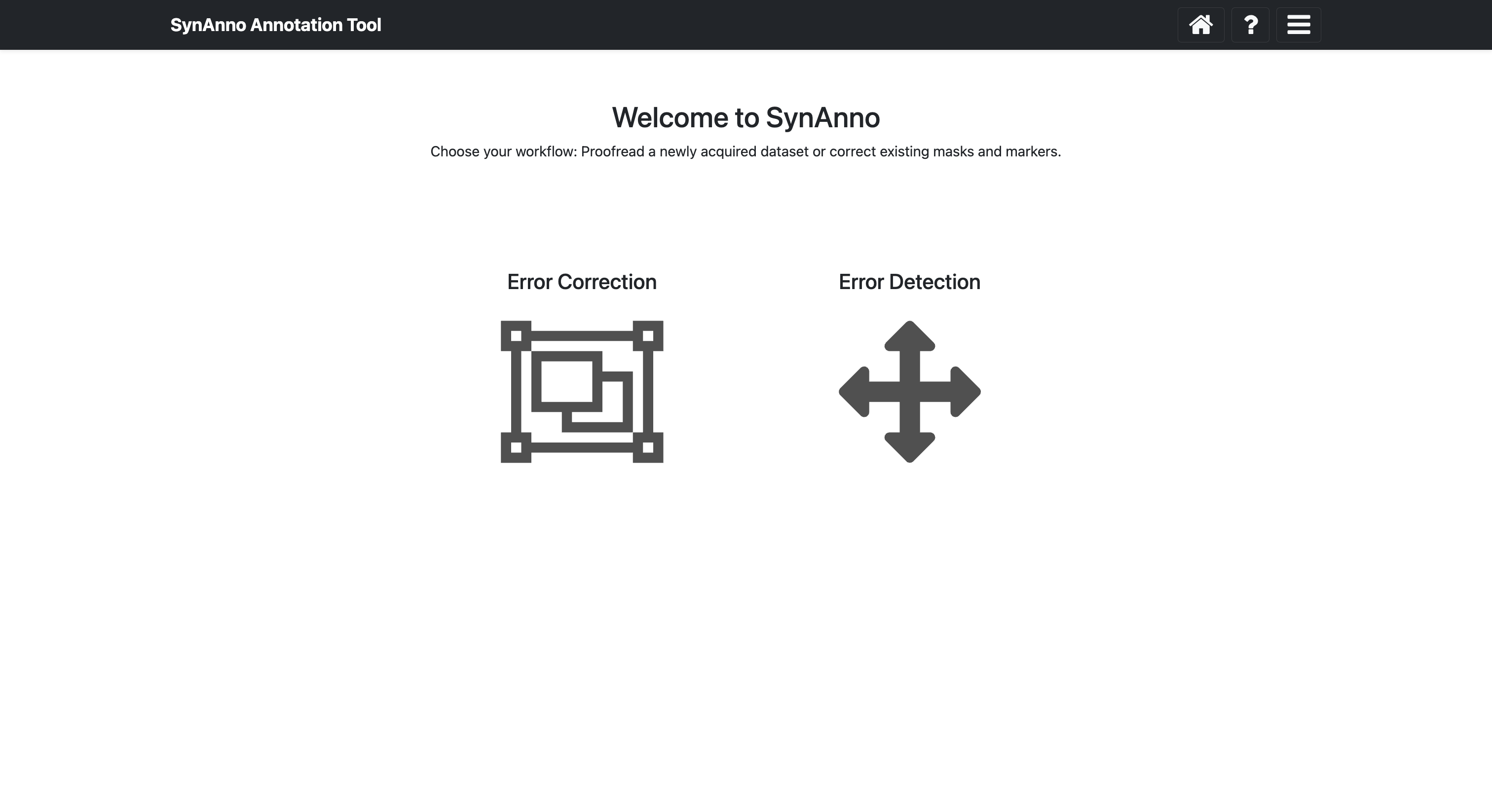
# the app is accessible at http://127.0.0.1/5000/On the landing page you can choose between two workflows: "Revise Dataset" and "Proofread Annotation". The former allows you to redraw segmentation masks, assign pre-/post-synaptic IDs, and add missed false negatives. The latter allows you to review existing segmentations and pre-/post-synaptic IDs, mark incorrect instances, and assign error descriptions to those instances. After categorizing all erroneous instances in the "Proofread Annotation" workflow, you can automatically proceed with the "Revise Dataset" workflow.
Each page has three buttons: "Home", "Question Mark", and "Menu". The first returns you to the landing page. The second provides an explanation of the current view and its functionality. The third provides general information and contact details.
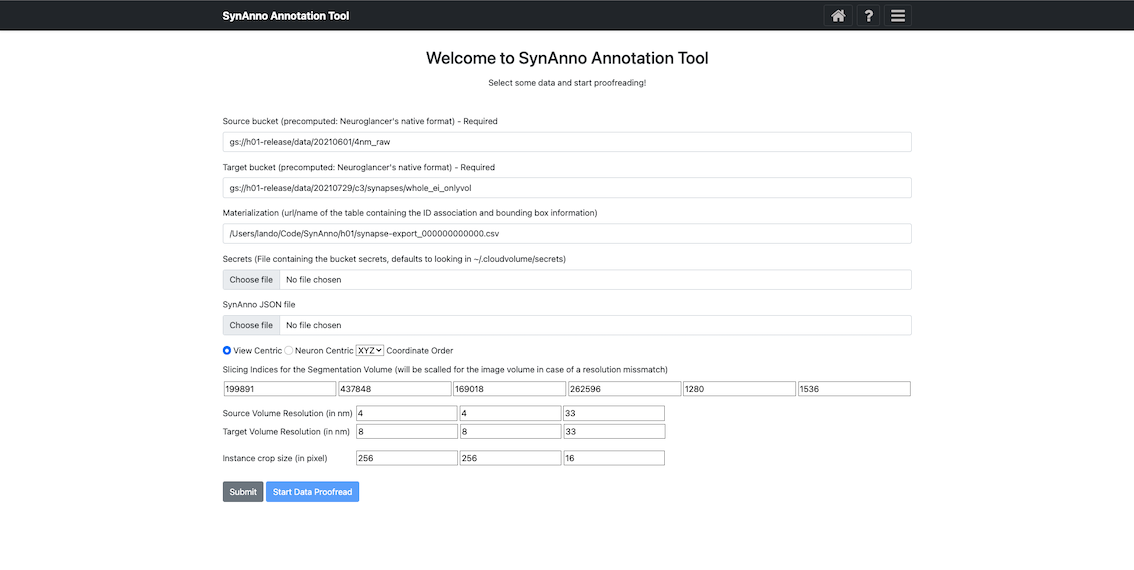
This view is identical for both workflows. You'll be prompted to provide a source bucket and target bucket (both in Neuroglancer's precomputed format), a URL to a materialization table, bucket secrets (defaults to ~/.cloudvolume/secrets), and optionally a JSON file containing instance metadata. The JSON file can be used to save and restore sessions or start a "Revise Dataset" workflow with information from a "Proofread Annotation" workflow.
You can load instances by querying the materialization table based on sub-volume constraints in the 'View Centric' approach or based on pre-/post-synaptic coordinates in the 'Neuron Centric' approach. You'll also need to specify the coordinate layout of the referenced precomputed datasets, the source volume resolution (in nm), the target volume resolution (in nm), and instance crop size (in pixels).
After providing the required information, click 'Submit' to prepare the data for the first page or revision. Then, click "Start Data Proofread"/"Start Drawing" to begin proofreading or revision.
- Workflow: Proofreading
- URL: http://127.0.0.1:5000/annotation
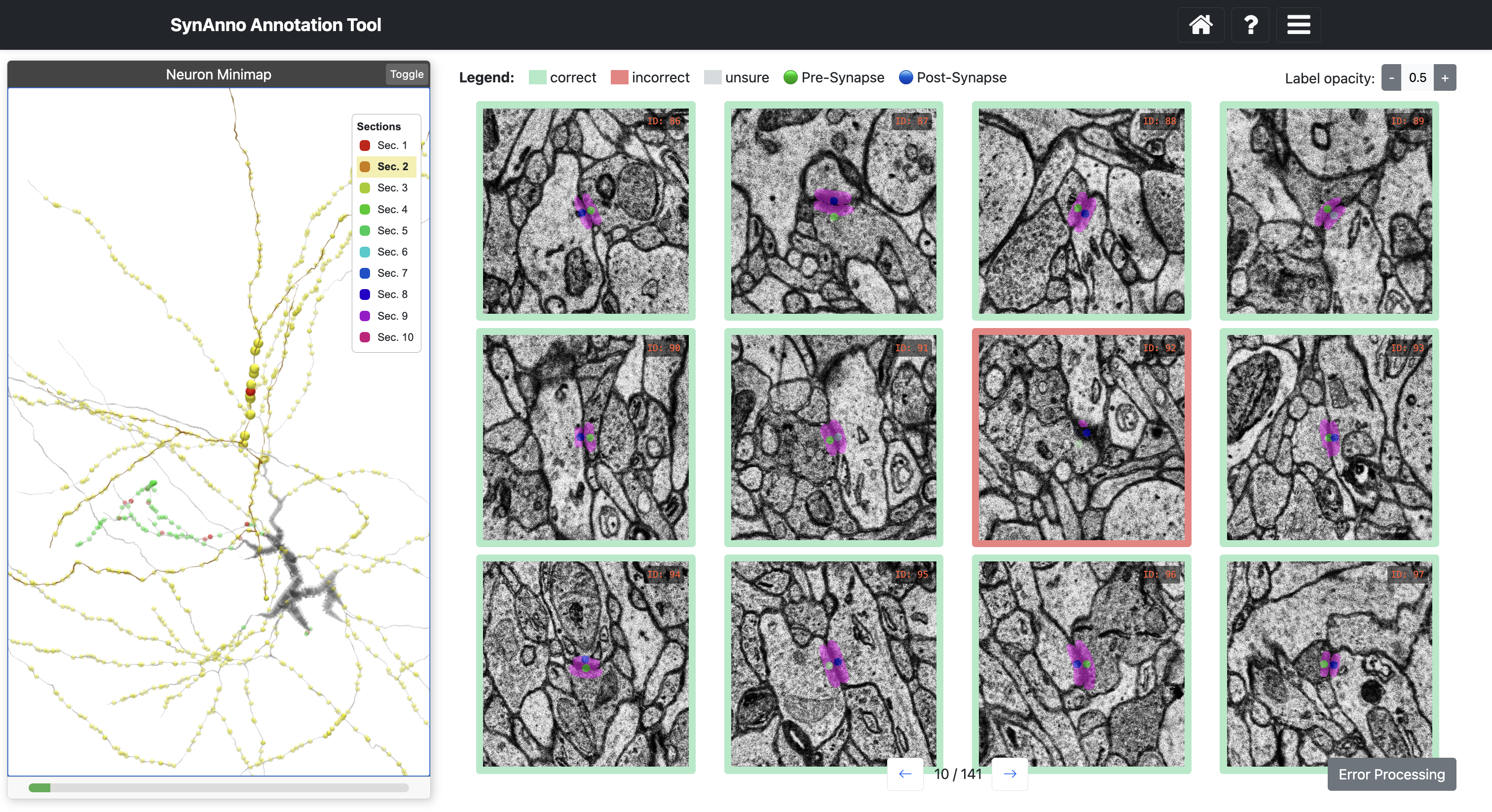
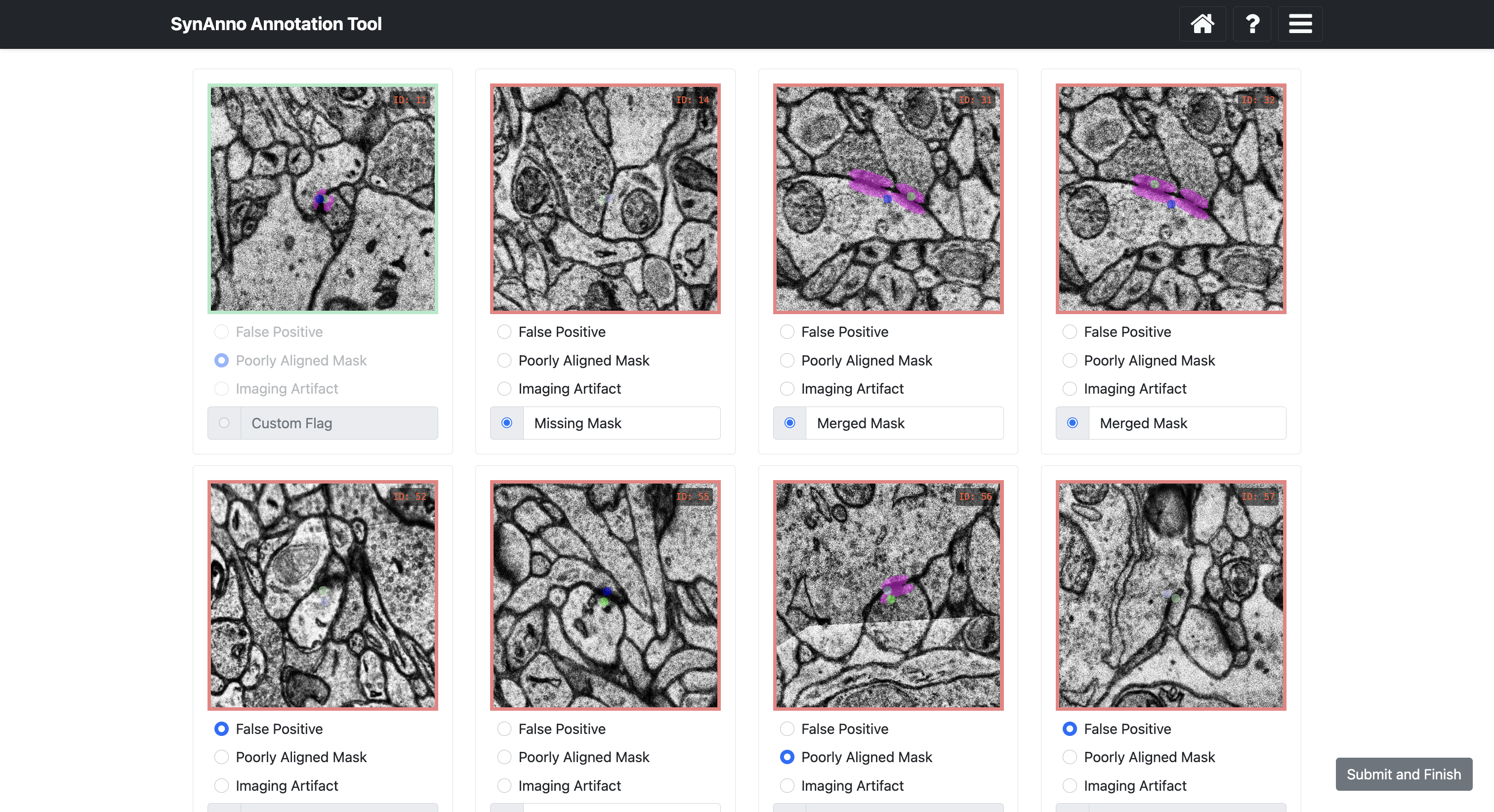
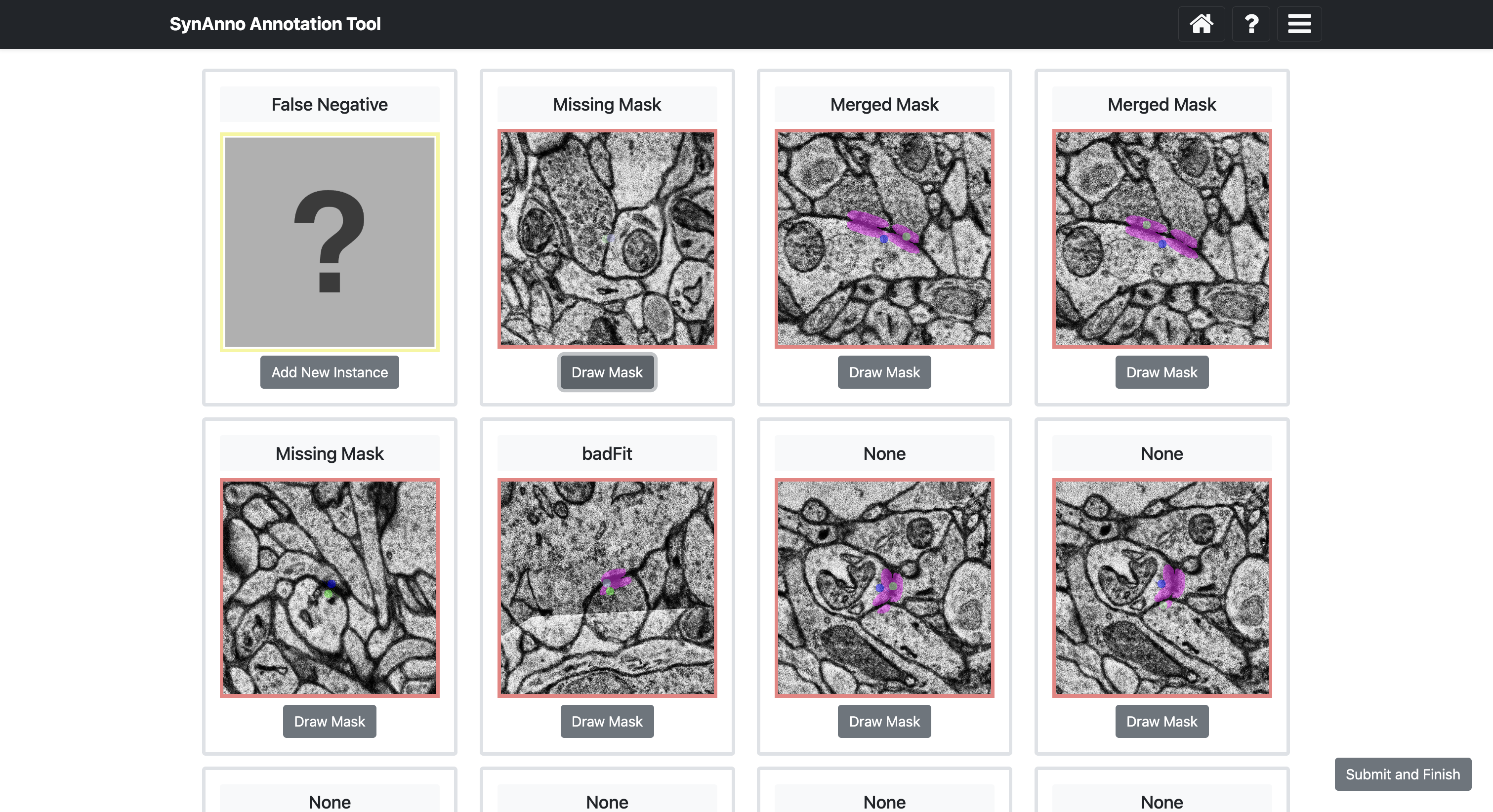
Clicking Start Data Proofread directs you to a grid view of instances. Instance status is indicated by color: correct (green), incorrect (red), unsure (gray). Clicking on an instance changes its status: once for incorrect, twice for unsure, three times for correct.
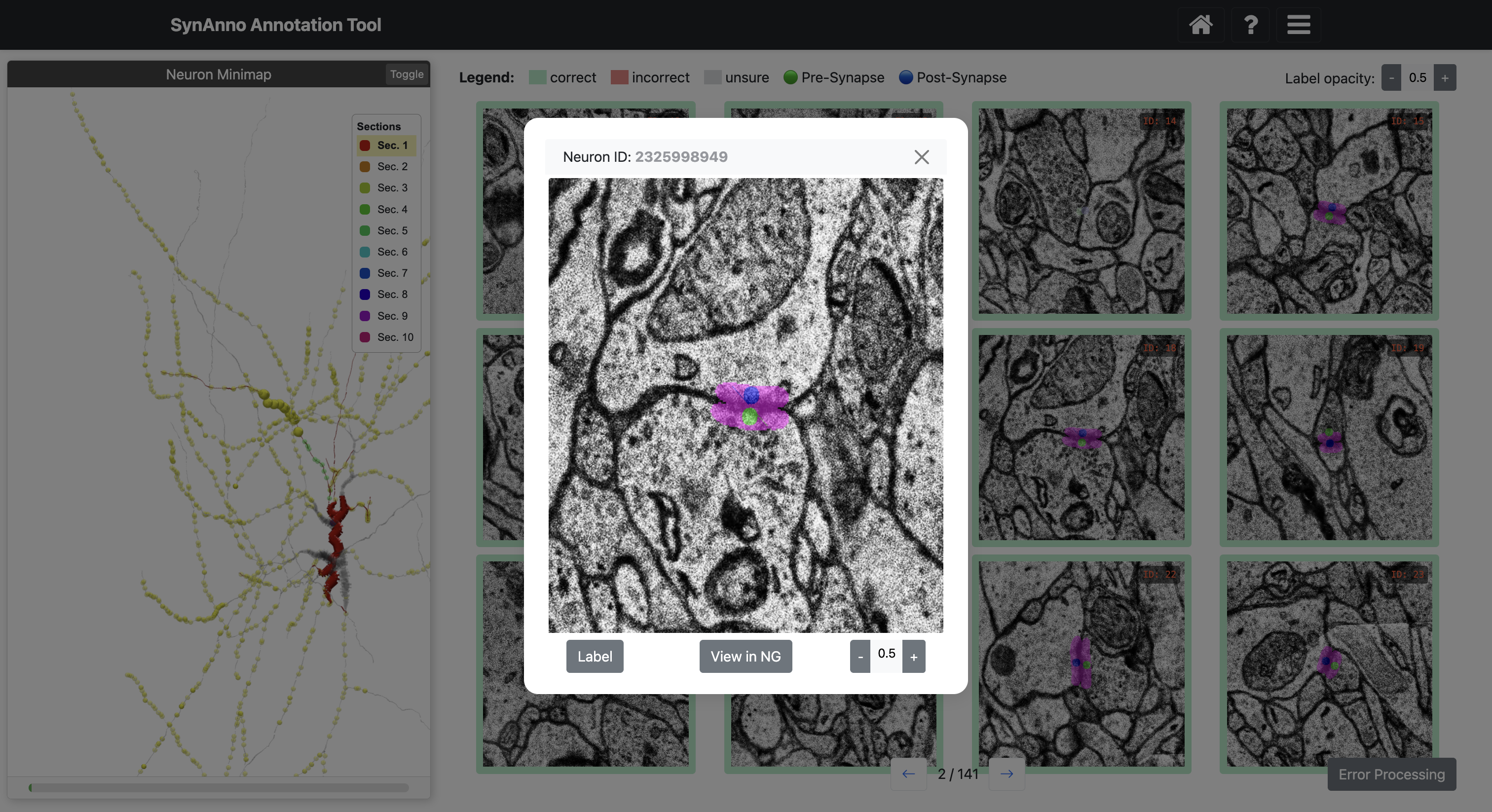
To inspect an instance's mask, right-click the instance to enlarge the patch and navigate through slices.
Click View in NG to view the instance in Neuroglancer.
After evaluating the segmentation masks, click -> to load and evaluate the page. When done, click Error Processing on the last proofreading page.
- Workflow: Proofreading
- URL: http://127.0.0.1:5000/categorize
Clicking Error Processing brings you to the error categorization view. Here, you specify errors for instances marked as incorrect or unsure. Scroll sideways to see all cards. Right-click to enlarge the patch, navigate through slices, or open Neuroglancer. When done, click Submit and Finish.
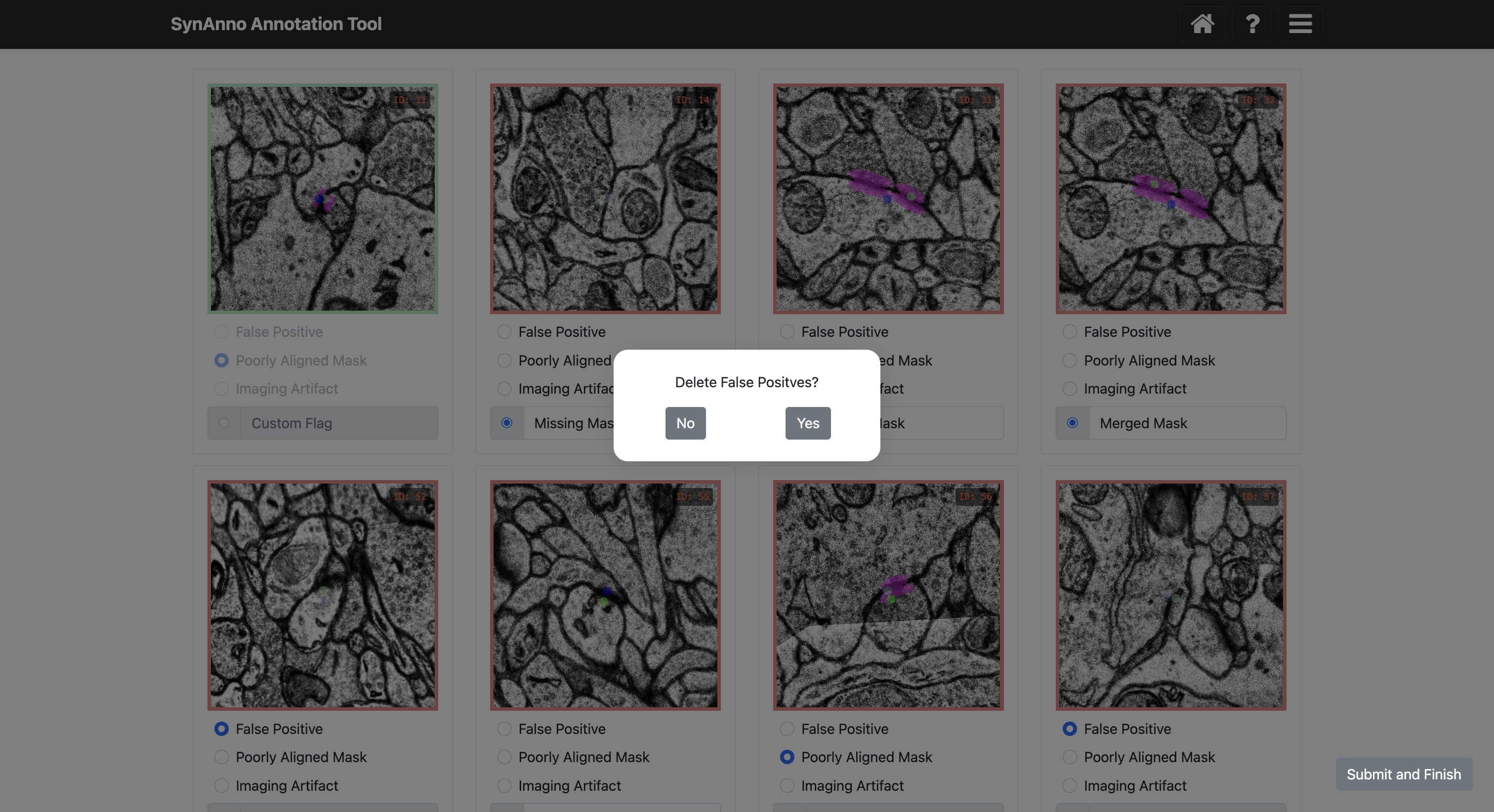
If you marked instances as false positives, you'll be asked if they should be discarded.
- Workflow: Proofreading
- URL: http://127.0.0.1:5000/export_annotate
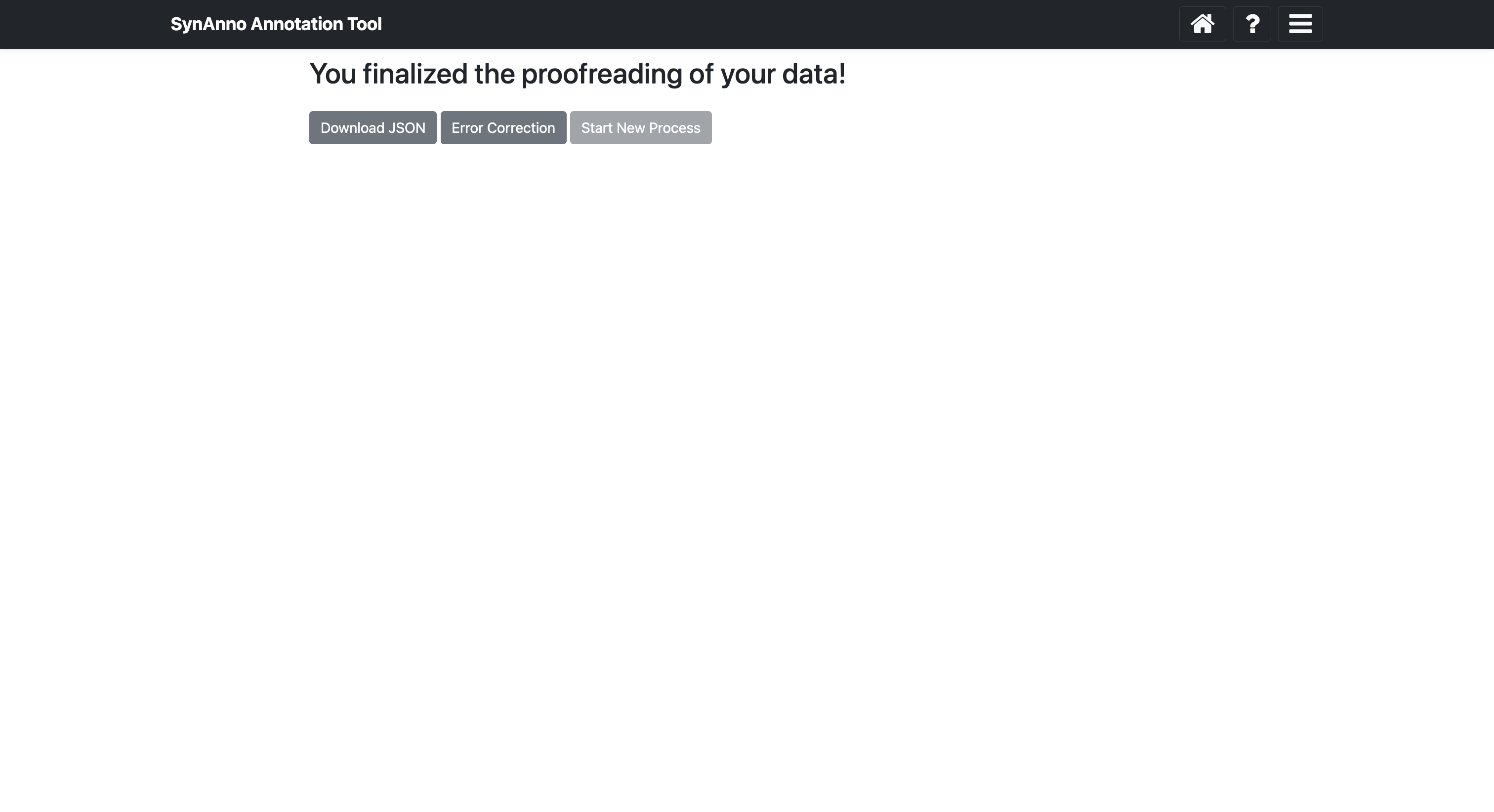
After clicking Submit and finish, you can download the JSON file containing instance metadata by clicking Download JSON, redraw masks with the Revise Dataset workflow by clicking Redraw Masks, or start a new process by clicking Start New Process.
- Revision Workflow
- http://127.0.0.1:5000/draw
Clicking Start Drawing from the Open Data view, after choosing the Revision Workflow on the Landing Page, or selecting Redraw Masks in the Export Annotations view, will take you to the Draw view. In this view, you can create segmentation masks, set pre-/post-synaptic IDs, and add missed false negatives. If you arrived here from the Proofreading Workflow, you'll see instances marked as incorrect or unsure during previous proofreading, along with their associated error labels assigned during categorization. If you arrived from the Revision Workflow, you'll see all instances associated with the given pre-/post-synaptic IDs or within the specified sub-volume range that have incorrect or unsure labels in the provided JSON file. If you don't provide the JSON file, you can only add missed false negatives, as the tool assumes an initial label of correct for all instances.
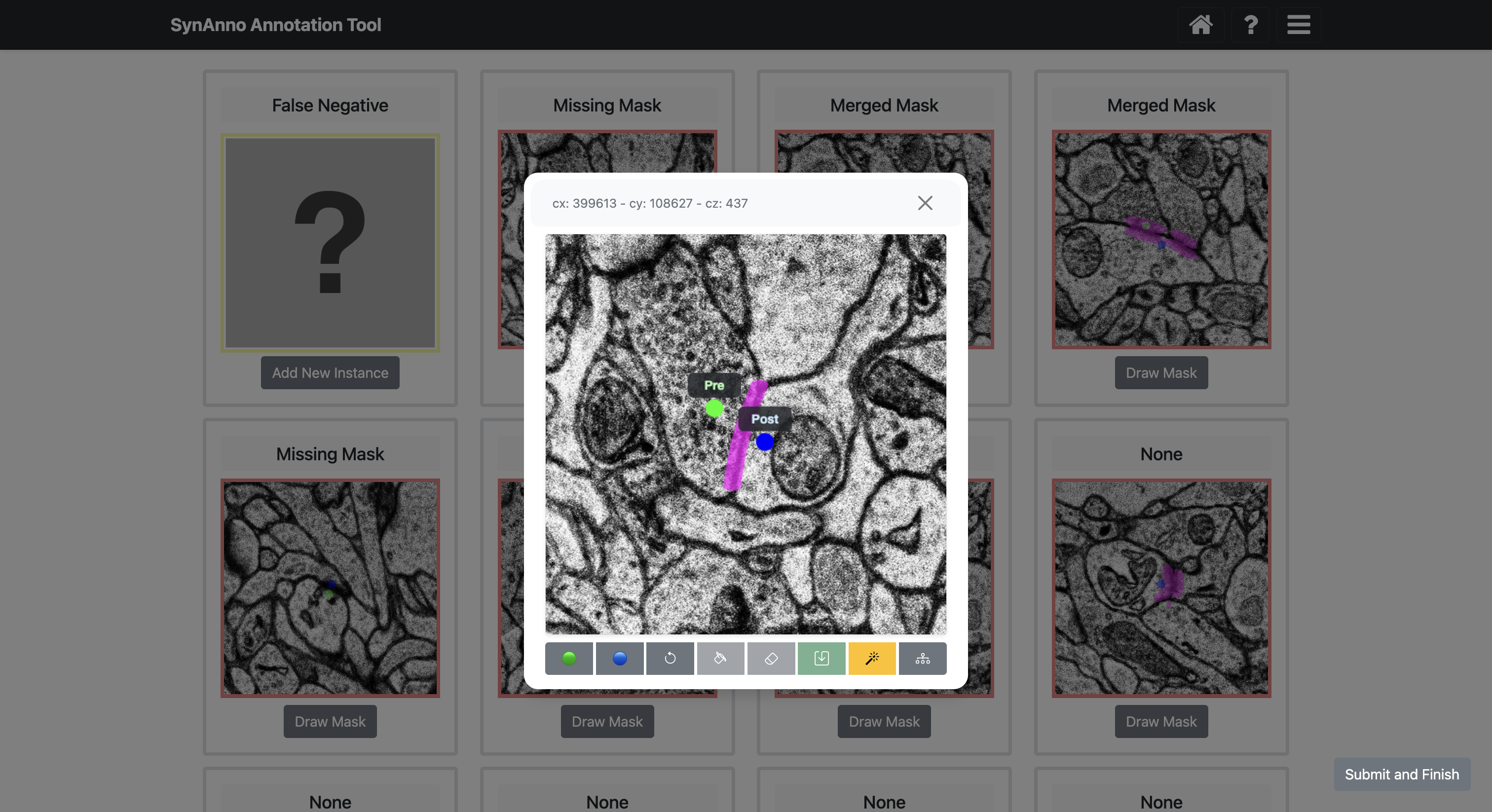
Selecting an instance and clicking Draw Mask will open a view specific to that instance. In this view, you can scroll through all slices of the instance, draw masks for as many slices as you like, and set the pre- and post-synaptic coordinate ID markers on any chosen slice. Clicking the Draw Mask button allows you to create a mask using spline interpolation with intuitive control points. After positioning all control points, click the Fill button to generate the mask. You can modify or erase parts of the drawn mask by clicking the Revise button, which acts like an eraser. Once satisfied with the mask, click Save to save it for that slice. To set the pre- and post-synaptic coordinate ID markers, select the appropriate slice, click the Pre-Synaptic CRD or Post-Synaptic CRD button, and then click the relevant coordinate location. At any time, you can click View in NG to open the instance in Neuroglancer for a better view of marker placement or mask drawing. Upon closing the instance view, you will see the slice and its custom mask displayed in the sideways-scrollable overview for which you drew the custom mask last.
To add previously missed false negatives, click the Add New Instance button to open Neuroglancer. Navigate to the location of the missed false negative, position your cursor at the relevant location, and press the c key on your keyboard to set a yellow marker. After marking the location, click the Review button to open a module displaying the chosen location's coordinates and the slice depth for the instance's bounding box. Confirm the settings by clicking Save, or manually adjust the values before saving. This adds the missed false negative to the list of instances available for segmentation mask drawing.
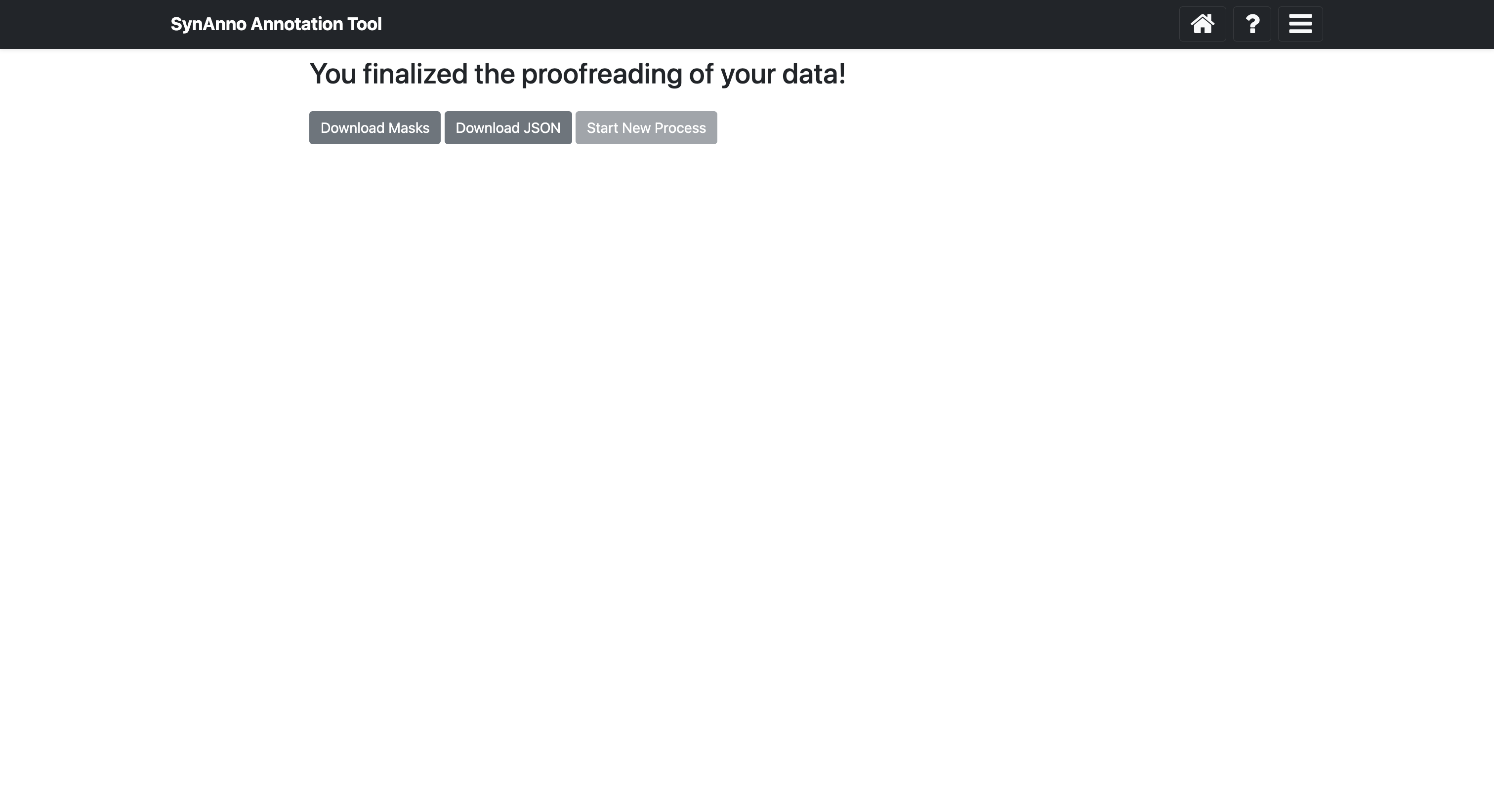
After completing the segmentation masks and setting the pre and post-synaptic coordinate ID markers for all instances, click the Submit and Finish button to proceed to the final view.
In this view, you can download the JSON file containing the instances' metadata by clicking Download JSON, or download the segmentation masks as a numpy array or image by clicking Save Masks. The instance ID, bounding box, and slice number are encoded in the file names. If you want to start a new process, click Start New Process to return to the landing page.
Pre-submission, we ask you to only create issues for bugs, feature requests, or questions about the code. After submission, we strongly encourage any kind of contribution, including bug fixes, additional features, and documentation improvements. If you're unsure about whether a contribution is appropriate, feel free to open an issue and ask.
For members of the VSC, please set up pre-commit hooks to ensure that your code adheres to our coding style. This will prevent you from committing code that doesn't follow our style guidelines and subsequent failing PRs. To set up pre-commit hooks, run the following command in the root of the SynAnno repository.
pre-commit installYou can manually run pre-commit on all the files in your repository by running:
pre-commit run --all-filesNow, whenever you try to commit changes to your repository, pre-commit will automatically run all hooks. If any problems are found, the commit will be prevented until you fix them.