Generative adversarial networks for single-cell RNA-seq imputation
Now published in Nucleic Acids Research, https://doi.org/10.1093/nar/gkaa506
scIGANs is a computational tool for single-cell RNA-seq imputation and denoise using Generative Adversarial Networks (GANs). Build on PyTorch, scIGNAs enables GPU acceleration inaddition to CPU computing as long as the GPUs are available.
scIGANs currently can only be built and run on Linux/Unix-based systems.
- Download
git clone https://github.com/xuyungang/scIGANs.git cd scIGANs- Install
bash scIGANs.installer [-p dir/to/install]- use
-poption to direct the installation to a sepecific directory; default, current working directory. - You may need to start a new terminal or
source ~/.bashrcto enablescGANsexecutable from your$PATH.
- use
- Check installation
scIGANs -h- For successful installation, you will see the help message.
- Occasionally, you may need to restart you terminal to run
scIGANs. - If you installed scIGANs before, please use
which scIGANsto check the currently active version.
scIGANs is implemented in python (>2.7) and R(>3.5). Please install Python (>2.7), R(>3.5) and all dependencies before run scIGANs. Users can either use pre-configured conda environment (recommended) or build your own environment manually.
- Install conda (>4.5)
- If you already got conda installed, go to step
2directly. - Downlowd bash installer (or find different version at https://docs.conda.io/en/latest/miniconda.html)
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
- Install conda (Linux for example)
bash miniconda/Miniconda3-latest-Linux-x86_64.sh- follow the prompted instruction to complete the installation.
- Create conda envirenment for scIGANs
-
cd scIGANsNote: if you have scIGNAs conda environment configured before, please remove it before proceeding.
conda env remove -n scIGNAs -
For Linux:
conda env create -f scIGANs.conda.env.Linux.yml -
For Mac OS:
conda env create -f scIGANs.conda.env.Mac.yml
- Activate conda envirenment for scIGANs
- Activate conda environment:
conda activate scIGNAs - Test scIGNAs
- R packages: scIGANs requires two R packages:
SamSPECTRALandRtsne. Make sure you have the permission to install R packages to the default lib. - python modules:
pytorch,numpy(will be installed with pytorch when usingconda install),pandas,torchvision, andjoblibare required for scIGANS.
-
Usage:
scIGAN in.matrix.txt [options] -
Options:
-h --helpShow this usage message and exit scIGANs.- Input:
in.matrix.txt[STR] A tab-delimited text file (including appropriate PATH), containing the expression counts with genes in rows and cells in columns. The first row is header and first column is gene IDs or names. <required>-l|--label_file[STR] A text file (including appropriate PATH) contains the labels (cell types, subpopulations, etc.), one label per line per cell with the same order in in.matrix.txt. <optional> Default: scIGANs will learn the subpopulations using Spectral clustering.
- Training:
-e|--epochs[INT] The number of epochs to train the GANs. <optional> Default: 200-d|--latent_dim[INT] Dimension of the latent space of generator. <optional> Default: 100-b|--batch_size[INT] The number of samples per batch to load.More samples per batch requires more memory.<optional> Default: 8. NOTE: no more than cell number.-j|--job_name[STR] A string will be used as prefix to name all output files.<optional> Default: prefix=<in.matrix.txt>-<label_file_name>-t|--threthold[FLOAT] Convergence threthold. <optional> Default: 0.01.
- Imputing:
--imputeSet this option to skip training and directly impute using pretrained model for the same input settings.-s|--sim_szie[INT] Number of generated datasets for imputing. <optional> Default: 200-k|--knn_n[INT] Number of nearest neighbours for imputing. <optional> Default: 10
- Output:
-o|--outdir[STR] Output directory. <optional> Default: current working directory
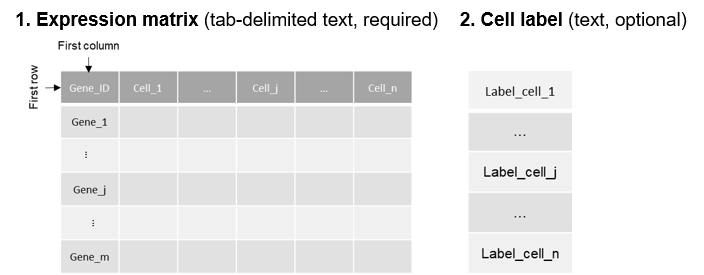
scIGANs takes tab-delimited text file(s) as input. The expression count matrix file is required and needs to be imputed. The cell labels file is optional and contains the cell labels, each per row with the same cell order as in expression matrix file. The following shows the toy example formats.
scIGANs will generate following files:
Model files:one file for Discriminator and one file for Generator are located in thhe folderGANs_models/. Files are named in the format of<in.matrix.txt>-<label_file_name>-<latent_dim>-<n_epochs>-<cluster_number>-g.ptor<in.matrix.txt>-<label_file_name>-<latent_dim>-<n_epochs>-<cluster_number>-d.pt.Imputed matrix:expression matrix, with the same format as input and only has some zero-counts replaced with expression values imputed by scIGANs. File is named in the format ofscIGANs_<timestamp>_<in_matrix_filename>.Log file:a log file of the running, named asscIGANs_<timestamp>_<in_matrix_filename>.log.
- Without label file:
scIGANs scIGNAs/install/path/test_data/ercc.txt -e 1 [options] - With label file:
scIGANs scIGNAs/install/path/test_data/ercc.txt -l ercc.label.txt -e 1 [options]
Yungang Xu yungang.xu@uth.tmc.edu
Zhigang Zhang zzg@hbue.edu.cn
Xiaobo Zhou xiaobo.zhou@uth.tmc.edu
Yungang Xu, Zhigang Zhang, Lei You, Jiajia Liu, Zhiwei Fan, Xiaobo Zhou, scIGANs: single-cell RNA-seq imputation using generative adversarial networks, Nucleic Acids Research, 2020, gkaa506, https://doi.org/10.1093/nar/gkaa506