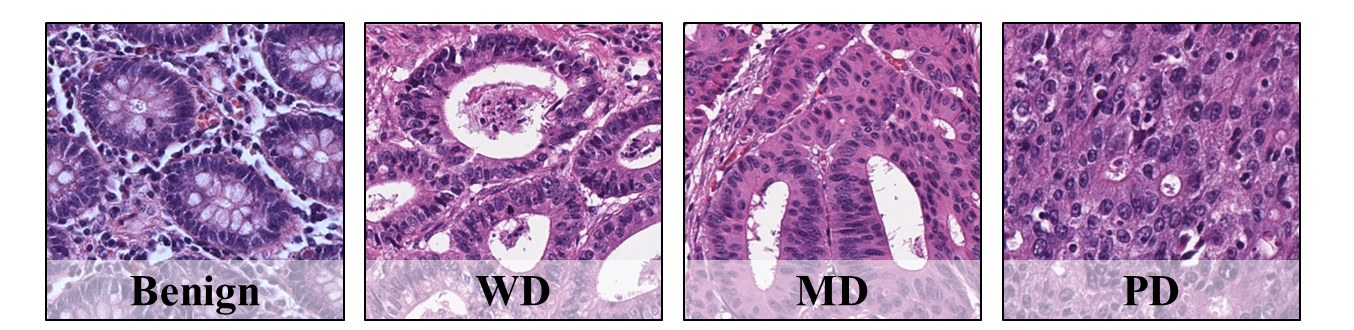
This repository provides the KBSMC colon cancer grading dataset that has been introduced in the paper:
Joint Categorical and Ordinal Learning for Cancer Grading in Pathology Images
Link to paper on Medical Image Analysis.
Google drive:
Training + validation + testing I: [link]
Testing II (Independent test set): [link]
The tissue images and annotations are provided by Kangbuk Samsung Hospital, Seoul, South Korea.
Two pathologists have delineated the annotation: Kim, Kyungeun, and Song, Boram.
Herein, we obtained the benign (BN) and three cancer ROIs, including well-differentiated (WD) tumor, moderately-differentiated (MD) tumor, and poorly-differentiated (PD) tumor.
-
The train+val+test sets contain 3 whole slide images (WSIs) and 6 colorectal tissue microarrays (TMAs) from 340 patients that were scanned at 40x magnification using an Aperio digital slide scanner (Leica Biosystems).
-
The Test_set_2 (Independent test set) includes 45 WSIs from 45 patients scanned using a NanoZoomer digital slide scanner (Hamamatsu Photonics K.K.), at 40x magnification.
-
The patch images are generated at 40x of size (~258 µm x 258 µm), then resize to 512x512 pixels (20x). For more detail, please refer to the paper above.
| Status | Training | Validation | Testing I | Testing II |
|---|---|---|---|---|
| Benign | 773 | 374 | 453 | 27986 |
| WD | 1866 | 264 | 192 | 8394 |
| MD | 2997 | 370 | 738 | 61985 |
| PD | 1397 | 234 | 205 | 11895 |
├── KBSMC_colon_tma_cancer_grading_512
│ ├── tma01
│ │ ├── 100100_19493_0.jpg
│ │ ├── 100221_64120_3.jpg
│ │ ├── 100383_42563_2.jpg
│ │ ├── 100443_52718_2.jpg
│ │ ├── 100634_14705_1.jpg
│ │ ├── ...
│ ├── tma02
│ ├── tma03
│ ├── tma04
│ ├── tma05
│ ├── tma06
│ ├── wsi01
│ ├── wsi02
│ └── wsi03
Class label is the last digit in image name (bolden), for example 100100_19493_0.jpg belongs to benign class.
Check out the dataset.py
def prepare_colon_tma_data(data_root_dir='./KBSMC_colon_tma_cancer_grading_512'):
""" List all the images and their labels
return Training + validation + testing I
"""
def load_data_info(pathname):
file_list = glob.glob(pathname)
label_list = [int(file_path.split('_')[-1].split('.')[0]) for file_path in file_list]
print(Counter(label_list))
return list(zip(file_list, label_list))
set_tma01 = load_data_info('%s/tma01/*.jpg' % data_root_dir)
set_tma02 = load_data_info('%s/tma02/*.jpg' % data_root_dir)
set_tma03 = load_data_info('%s/tma03/*.jpg' % data_root_dir)
set_tma04 = load_data_info('%s/tma04/*.jpg' % data_root_dir)
set_tma05 = load_data_info('%s/tma05/*.jpg' % data_root_dir)
set_tma06 = load_data_info('%s/tma06/*.jpg' % data_root_dir)
set_wsi01 = load_data_info('%s/wsi01/*.jpg' % data_root_dir) # benign exclusively
set_wsi02 = load_data_info('%s/wsi02/*.jpg' % data_root_dir) # benign exclusively
set_wsi03 = load_data_info('%s/wsi03/*.jpg' % data_root_dir) # benign exclusively
train_set = set_tma01 + set_tma02 + set_tma03 + set_tma05 + set_wsi01
valid_set = set_tma06 + set_wsi03
test_set = set_tma04 + set_wsi02
return train_set, valid_set, test_set
def prepare_colon_wsi_data(data_root_dir='./KBSMC_colon_45wsis_cancer_grading_512 (Test 2)'):
""" List all the images and their labels
return test_set 2
"""
def load_data_info_from_list(data_dir, path_list):
file_list = []
for WSI_name in path_list:
pathname = glob.glob(f'{data_dir}/{WSI_name}/*/*.png')
file_list.extend(pathname)
label_list = [int(file_path.split('_')[-1].split('.')[0]) -1 for file_path in file_list]
print(Counter(label_list))
list_out = list(zip(file_list, label_list))
return list_out
wsi_list = ['wsi_001', 'wsi_002', 'wsi_003', 'wsi_004', 'wsi_005', 'wsi_006', 'wsi_007', 'wsi_008', 'wsi_009',
'wsi_010', 'wsi_011', 'wsi_012', 'wsi_013', 'wsi_014', 'wsi_015', 'wsi_016', 'wsi_017', 'wsi_018',
'wsi_019', 'wsi_020', 'wsi_021', 'wsi_022', 'wsi_023', 'wsi_024', 'wsi_025', 'wsi_026', 'wsi_027',
'wsi_028', 'wsi_029', 'wsi_030', 'wsi_031', 'wsi_032', 'wsi_033', 'wsi_034', 'wsi_035', 'wsi_090',
'wsi_092', 'wsi_093', 'wsi_094', 'wsi_095', 'wsi_096', 'wsi_097', 'wsi_098', 'wsi_099', 'wsi_100']
test_set = load_data_info_from_list(data_root_dir, wsi_list)
return test_set
class DatasetSerial(data.Dataset):
"""get image by index
"""
def __init__(self, pair_list, img_transform=None, target_transform=None, two_crop=False):
self.pair_list = pair_list
self.img_transform = img_transform
self.target_transform = target_transform
self.num = self.__len__()
def __getitem__(self, index):
"""
Args:
index (int): index
Returns:
tuple: (image, index, ...)
"""
path, target = self.pair_list[index]
image = pil_loader(path)
# # image
if self.img_transform is not None:
img = self.img_transform(image)
else:
img = image
return img, targetIf any part of this dataset is used, please give appropriate citation to our paper.
@article{le2021joint,
title={Joint categorical and ordinal learning for cancer grading in pathology images},
author={Le Vuong, Trinh Thi and Kim, Kyungeun and Song, Boram and Kwak, Jin Tae},
journal={Medical image analysis},
pages={102206},
year={2021},
publisher={Elsevier}
}