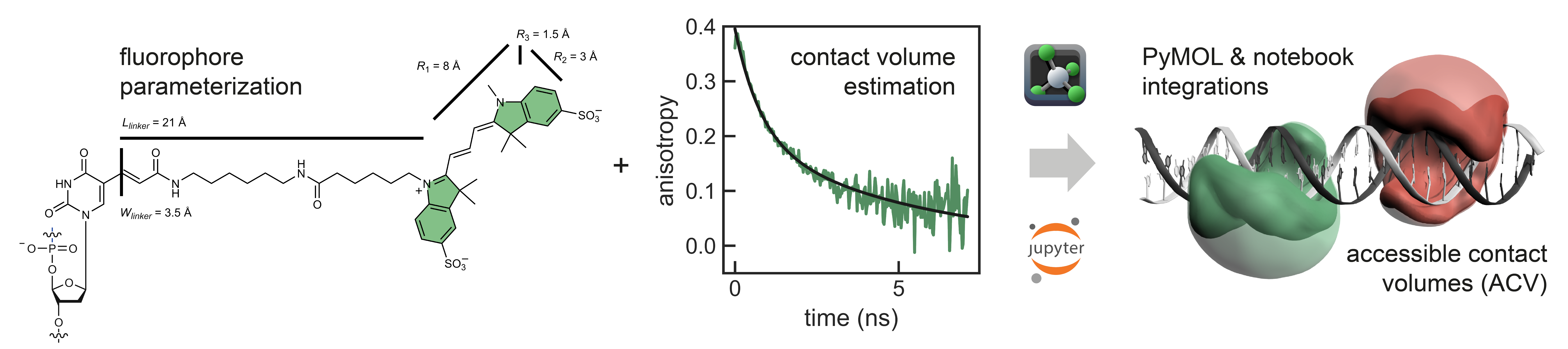
FRETraj is a Python module for predicting FRET efficiencies by calculating multiple accessible-contact volumes (multi-ACV) to estimate donor and acceptor dye dynamics. The package features a user-friendly PyMOL plugin1 for for FRET-assisted, integrative structural modeling. It interfaces with the LabelLib library for fast computation of ACVs. Specifically, FRETraj is designed to:
- plan (single-molecule) FRET experiments by optimizing labeling positions
- interpret FRET-based distance measurements on a biomolecule
- integrate FRET experiments with molecular dynamics simulations
Follow the instructions for your platform here
If you use FRETraj in your work please refer to the following paper:
- F.D. Steffen, R.K.O. Sigel, R. Börner, Phys. Chem. Chem. Phys. 2016, 18, 29045-29055.
- S. Kalinin, T. Peulen, C.A.M. Seidel et al. Nat. Methods, 2012, 9, 1218-1225.
- T. Eilert, M. Beckers, F. Drechsler, J. Michaelis, Comput. Phys. Commun., 2017, 219, 377–389.
- M. Dimura, T. Peulen, C.A.M. Seidel et al. Curr. Opin. Struct. Biol. 2016, 40, 163-185.
- M. Dimura, T. Peulen, C.A.M Seidel et al. Nat. Commun. 2020, 11, 5394.
1 PyMOL was developed by WarrenDeLano and is maintained by Schrödinger, LLC.