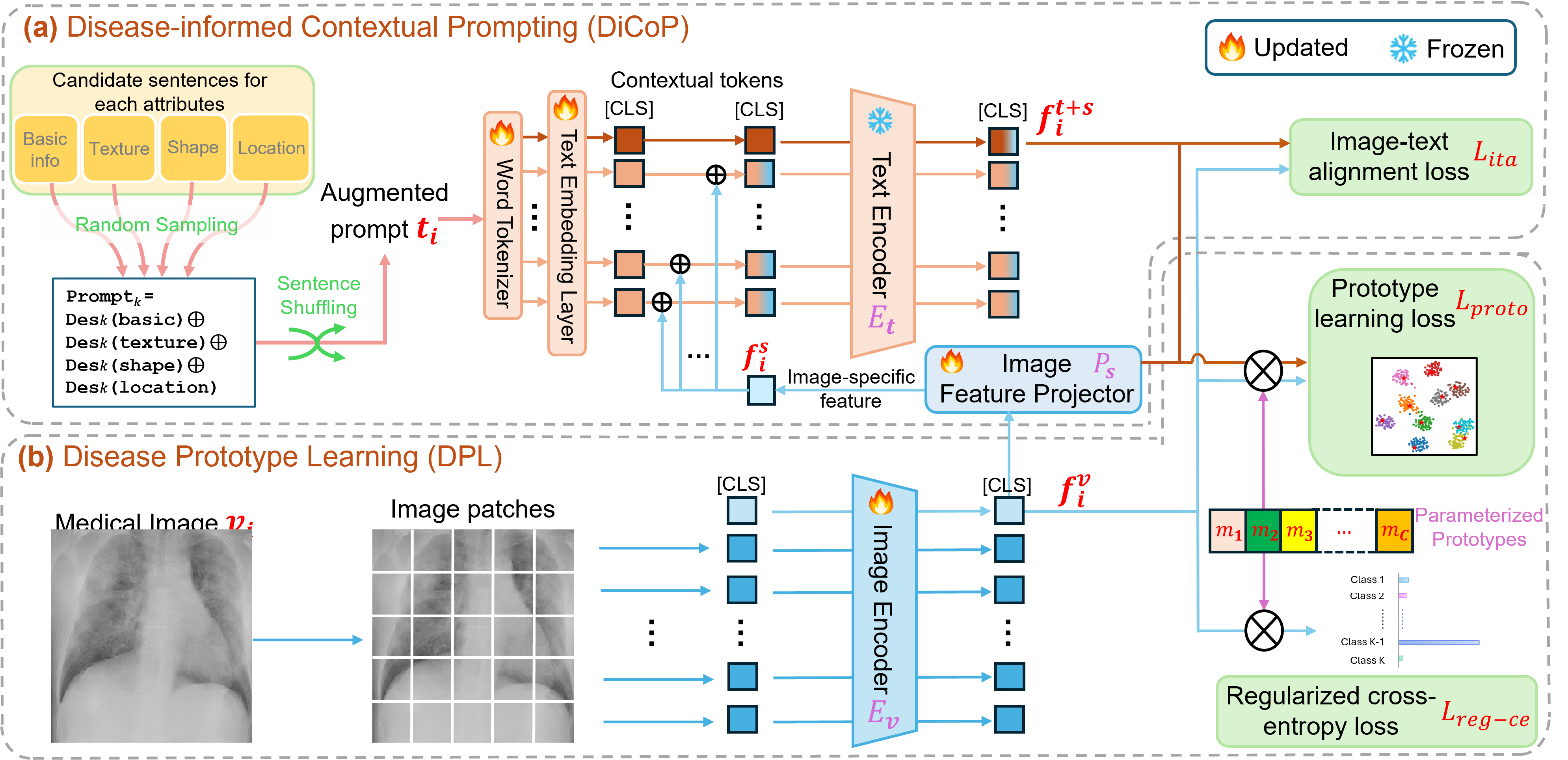
This repository includes extra detailed information of our Disease-informed Adaptation for adapting pre-trained medical Vision-Language Models (VLMs) to newly identified and/or under represented diseases.
The first version of our work is early accepted by MICCAI 2024.
More results and the source code will be released soon!
The complete set of prompt candidates for each attribute of every medical finding category is listed in the table below. Based on the disease-informed prompts (the Prompt candidate 1 column) from our radiologist, we also utilized GPT-4 to automatically generated the other two candidates (the Prompt candidate 2 and Prompt candidate 3 colums) for each descriptive attribute (texture, shape, and location) based on our predefined template. All these prompts were then manually revised with the help of a radiologist, ensuring that they are medically accurate.
| Medical findings (Categories) | Attributes | Prompt candidate 1 | Prompt candidate 2 | Prompt candidate 3 |
|---|---|---|---|---|
| COVID-19 pneumonia | Desk=1(basic)[1] | "A chest X-ray image of a patient with COVID-19." | "A radiograph of a COVID-19 patient." | "An X-ray image showing a patient diagnosed with COVID-19." |
| Desk=1(texture) | "Texture Patterns include bilateral, patchy and ground-glass opacities (GGO) in the lungs. These opacities can vary in density and distribution." | "Texture patterns feature bilateral, patchy and ground-glass opacities in the lungs, which may differ in density and distribution." | "Texture patterns exhibit bilateral, patchy and ground-glass opacities (GGO) in the lungs, varying in density and distribution." | |
| Desk=1(shape) | "The opacities can have irregular shapes, appearing as hazy areas with fuzzy borders." | "The opacities may exhibit irregular contours, manifesting as hazy regions with indistinct edges." | "The opacities often feature irregular forms, presenting as blurred areas with indeterminate boundaries." | |
| Desk=1(location) | "The opacities are commonly located in the peripheral regions of the lungs, particularly in the lower lobes. They may involve multiple lung segments of both chest sides." | "The opacities typically appear in the peripheral areas of the lungs, especially in the lower lobes, and can affect multiple segments of both sides of the chest." | "Opacities are often found in the peripheral parts of the lungs, mainly within the lower lobes, and may affect several lung segments on both sides of the chest." | |
| Non-COVID-19 pneumonia | Desk=2(basic)[1] | "A chest X-ray image of a patient with pneumonia." | "A radiograph displaying the lung condition of a patient diagnosed with pneumonia." | "An X-ray image of a pneumonia patient." |
| Desk=2(texture) | "Textual Patterns can include areas of increased lung density due to inflammatory infiltrates." | "Textual patterns may feature regions of heightened lung density resulting from inflammatory infiltrates." | "Textural patterns can display areas of elevated lung density caused by inflammatory infiltrates." | |
| Desk=2(shape) | "In non-COVID pneumonia, opacities may have a lobar or segmental distribution, depending on the type of pneumonia." | "Opacities can present with a lobar or segmental distribution, varying according to the specific type of pneumonia." | "The distribution of opacities can be either lobar or segmental, based on the type of non-COVID pneumonia." | |
| Desk=2(location) | "The location of pneumonia opacities can vary but is often seen in specific lobes or segments of the lung." | "The position of pneumonia opacities varies but is usually observed in particular lobes or segments of the lung." | "Pneumonia opacities can appear in various locations but commonly manifest in specific lobes or segments of the lung." | |
| Healthy individuals | Desk=3(basic)[1] | "A chest X-ray image of a normal healthy individual." | "A chest X-ray showing the lungs of a normal, healthy individual." | "An X-ray image of the chest from a healthy individual" |
| Desk=3(texture) | "No respiratory symptoms or underlying lung conditions, chest typically show clear lung fields with no areas of abnormal opacities." | "In the absence of respiratory symptoms or pre-existing lung conditions, a chest X-ray generally reveals clear lung fields free from any abnormal opacities." | "Without respiratory symptoms or pre-existing lung conditions, a chest X-ray typically shows clear lung fields without any abnormal opacities." | |
| Desk=3(shape) | "No hazy areas with fuzzy borders." | "There are no unclear regions with blurred boundaries." | "There are no indistinct areas with blurred edges." | |
| Desk=3(location) | "The whole lung fields appear homogeneous and translucent without any irregularities or opacities." | "The entire lungs seem uniform and translucent, devoid of any irregularities or areas of opacity." | "The whole lung fields present as homogeneous and translucent, lacking any irregularities or opacities." |
[1] The default prompt template setting in the previous work, such as MaPLe and KgCoOp.
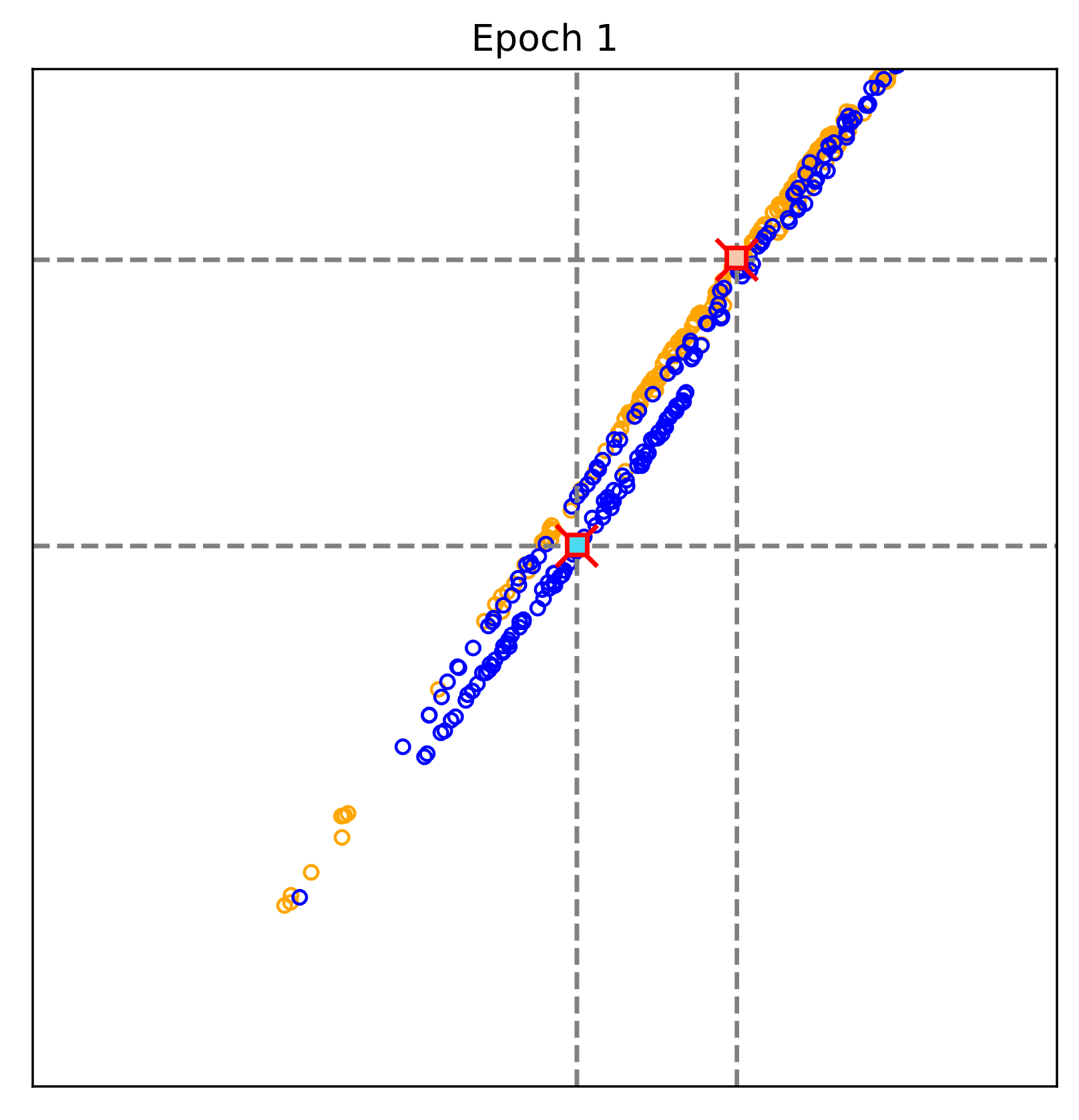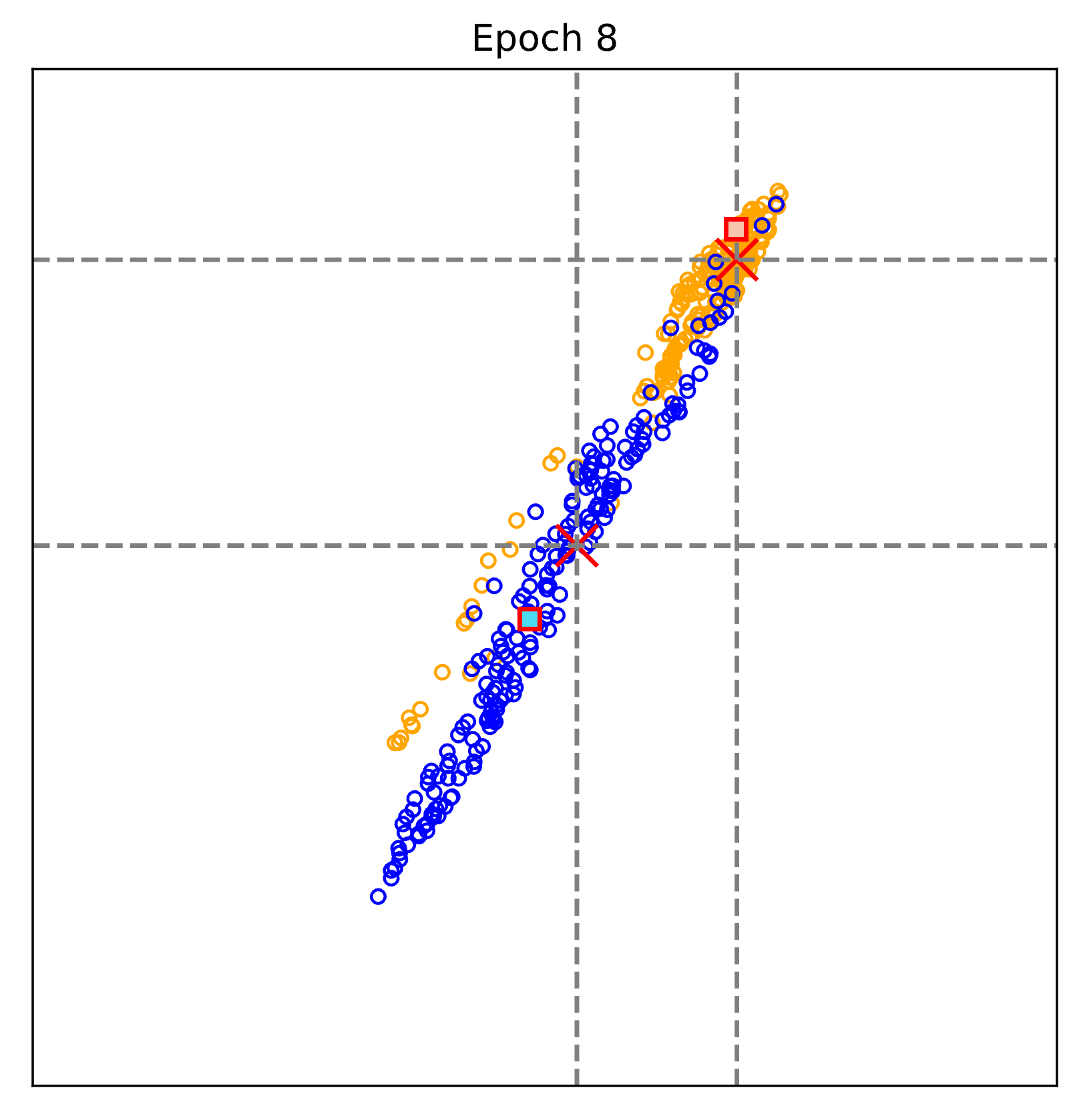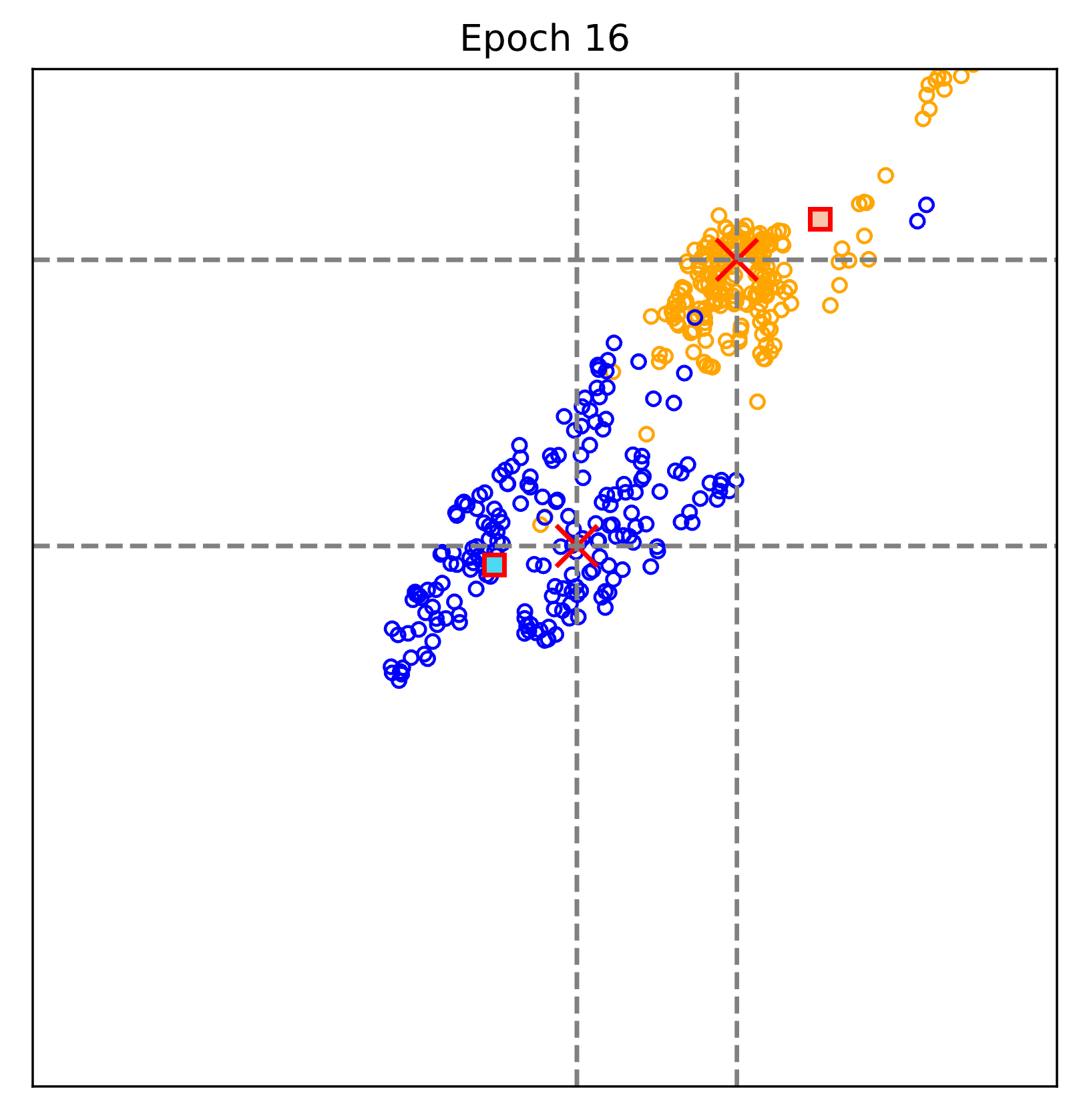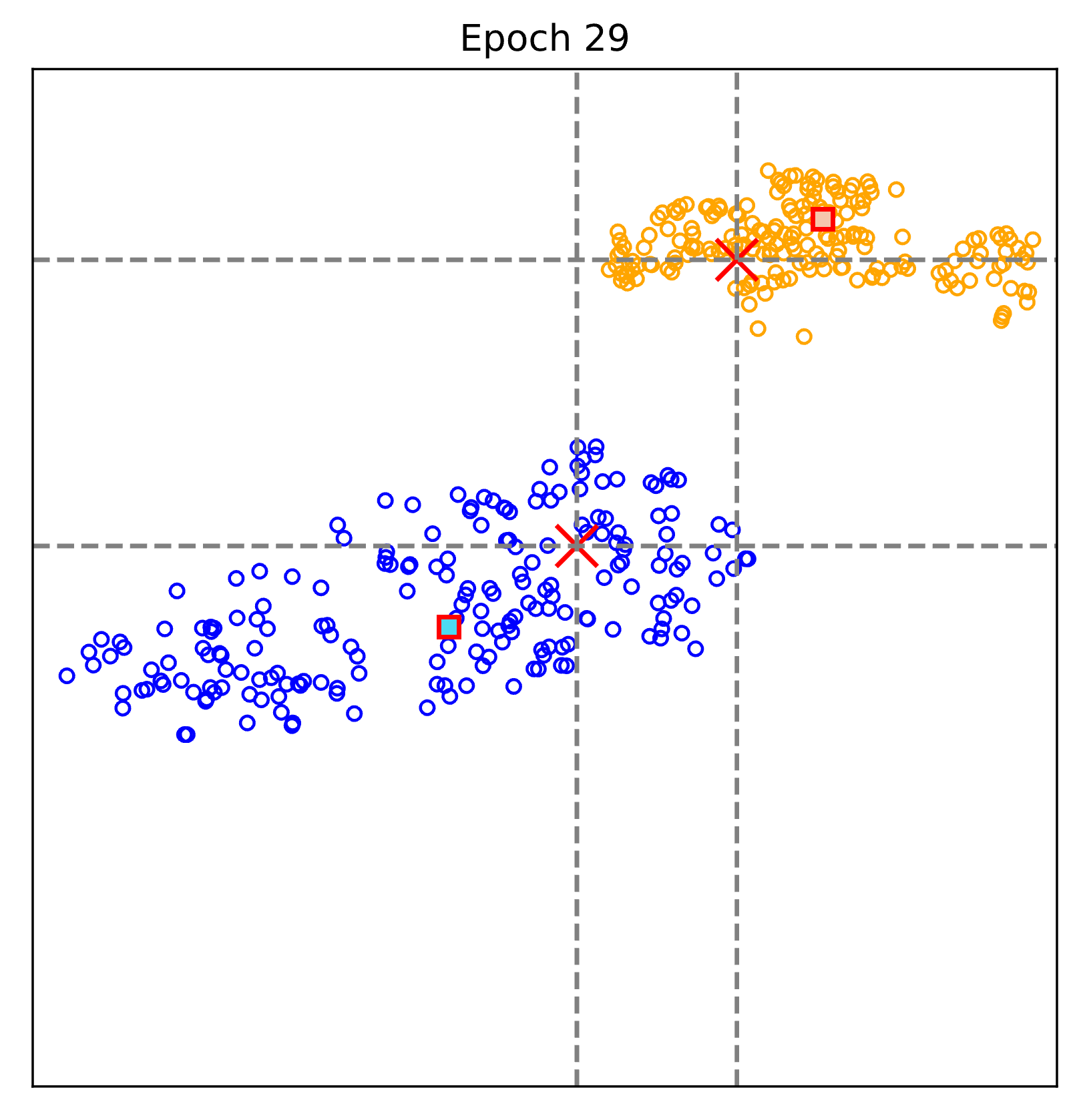
The figure above visualizes the evalution of the learnt representations epoch by epoch.
Blue: Non-COVID pneumonia samples; Orange: COVID-19 samples.
Red cross: text-defined prompts; rectangular (blue and orange): learnt prototypes of Non-COVID pneumonia and COVID-19.
- Python 3.9
- PyTorch 2.3.0+cu121
- HiggingFace transformers 4.18.0
- peft 0.7.1
- A computing device with GPU (>20G)
The two chest X-ray image datasets used in our work are publicly available.
The COVID-x datasets can be found here This dataset is already preprocessed by the challenge.
The COVID-sev dataset is can be found here. We follow the this work to preprocess the datasets.
Source code to be released ...
Source code to be released ...
Please cite these papers in your publications if it helps your research:
@article{zhang2024disease,
title={Disease-informed Adaptation of Vision-Language Models},
author={Zhang, Jiajin and Wang, Ge and Kalra, Mannudeep K and Yan, Pingkun},
journal={arXiv preprint arXiv:2405.15728},
year={2024}
}We would like to thank the authors we cited in our paper for sharing their codes.